Tuesday Poster Session
Category: Liver
P5867 - A Multi-Stage Machine-Learning Pipeline for Early Detection, Severity Stratification, and Prognostic Forecasting in Liver Disease
Tuesday, October 28, 2025
10:30 AM - 4:00 PM PDT
Location: Exhibit Hall

Sri Harsha Boppana, MBBS, MD
Nassau University Medical Center
Hicksville, NY
Presenting Author(s)
Sri Harsha Boppana, MBBS, MD1, Manaswitha Thota, MD2, Gautam Maddineni, MD3, Sachin Sravan Kumar Komati, 4, Sarath Chandra Ponnada, 5, Sai Lakshmi Prasanna Komati, MBBS6, C. David Mintz, MD, PhD7
1Nassau University Medical Center, East Meadow, NY; 2Virginia Commonwealth University, Richmond, VA; 3Florida State University, Cape Coral, FL; 4Florida International University, Florida, FL; 5Great Eastern Medical School and Hospital, Srikakulam, Srikakulam, Andhra Pradesh, India; 6Government Medical College, Ongole, Ongole, Andhra Pradesh, India; 7Johns Hopkins University School of Medicine, Baltimore, MD
Introduction: Liver disease remains a major global health burden, often progressing undetected until advanced stages. We applied supervised and unsupervised machine-learning techniques—including random forest classification, k-means clustering, and long short-term memory networks.
Methods: We analyzed the Indian Liver Patient Dataset (583 records) containing age, gender, total and direct bilirubin, alkaline phosphatase, alanine and aspartate aminotransferases, total proteins, albumin, and albumin–globulin ratio. We imputed missing albumin–globulin values with the mean, encoded gender numerically, and created interaction features (age × bilirubin, gender × alkaline phosphatase). We normalized all continuous variables and split the data into 80 percent training and 20 percent testing sets. A random forest classifier distinguished disease-positive from disease-negative cases. To infer severity without predefined labels, we applied k-means clustering (k = 3) on the normalized feature set and mapped each cluster to mild, moderate, or severe stages. We trained a second random forest on these inferred labels to predict severity for new patients. Finally, we generated synthetic five-visit time-series data—spacing visits 30 days apart and adding Gaussian noise—and trained long short-term memory networks to forecast next-visit bilirubin values and severity stages based on the preceding three visits.
Results: The disease classifier reached 81.2 % accuracy on the held-out test set, with precision and recall both above 0.80 for the liver-disease class. Clustering yielded three subgroups of comparable size, each exhibiting distinct biochemical signatures. The severity classifier achieved 99.1 % accuracy in assigning mild, moderate, or severe labels. The bilirubin-forecasting LSTM produced a mean absolute error of 1.07 mg/dL on the test set. The severity-forecasting LSTM achieved 96 % overall accuracy and an F1-score of 0.92 for the severe category. These results demonstrate that combining machine-learning classification, unsupervised clustering, and sequence modeling can accurately detect liver disease, stratify its severity, and anticipate biomarker and stage progression.
Discussion: The results demonstrate the potential of combining classification, clustering, and sequence modeling to support timely diagnosis, personalized risk stratification, and proactive management in liver disease care.
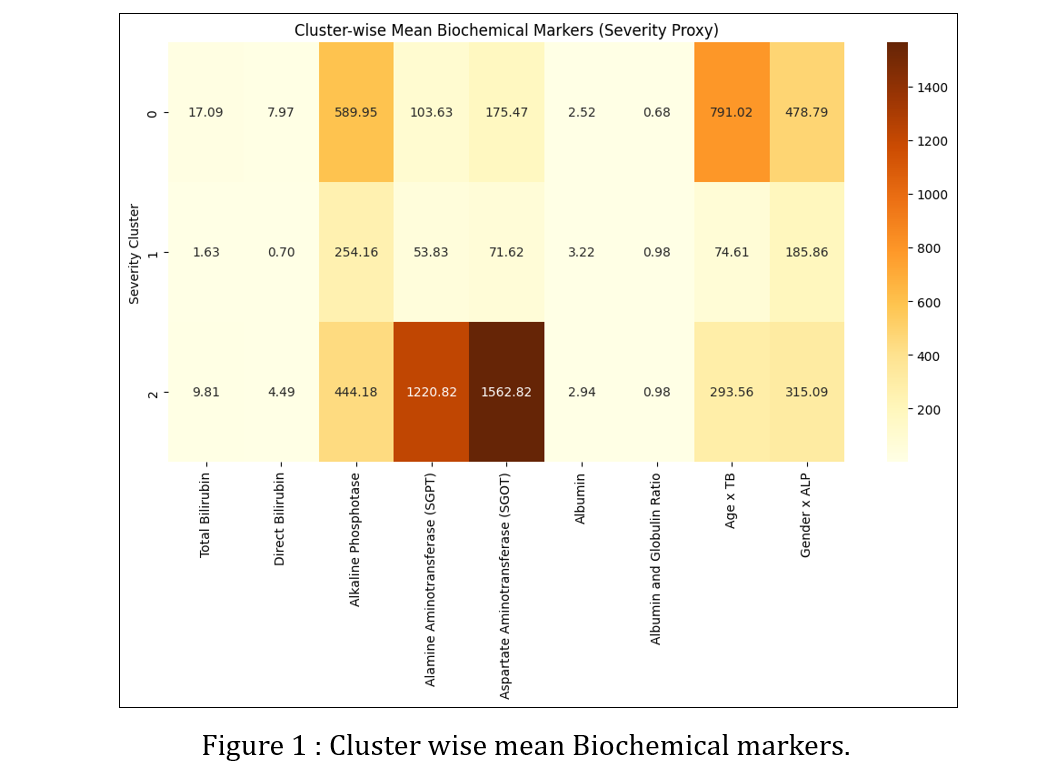
Figure: Figure 1
Disclosures:
Sri Harsha Boppana indicated no relevant financial relationships.
Manaswitha Thota indicated no relevant financial relationships.
Gautam Maddineni indicated no relevant financial relationships.
Sachin Sravan Kumar Komati indicated no relevant financial relationships.
Sarath Chandra Ponnada indicated no relevant financial relationships.
Sai Lakshmi Prasanna Komati indicated no relevant financial relationships.
C. David Mintz indicated no relevant financial relationships.
Sri Harsha Boppana, MBBS, MD1, Manaswitha Thota, MD2, Gautam Maddineni, MD3, Sachin Sravan Kumar Komati, 4, Sarath Chandra Ponnada, 5, Sai Lakshmi Prasanna Komati, MBBS6, C. David Mintz, MD, PhD7. P5867 - A Multi-Stage Machine-Learning Pipeline for Early Detection, Severity Stratification, and Prognostic Forecasting in Liver Disease, ACG 2025 Annual Scientific Meeting Abstracts. Phoenix, AZ: American College of Gastroenterology.
1Nassau University Medical Center, East Meadow, NY; 2Virginia Commonwealth University, Richmond, VA; 3Florida State University, Cape Coral, FL; 4Florida International University, Florida, FL; 5Great Eastern Medical School and Hospital, Srikakulam, Srikakulam, Andhra Pradesh, India; 6Government Medical College, Ongole, Ongole, Andhra Pradesh, India; 7Johns Hopkins University School of Medicine, Baltimore, MD
Introduction: Liver disease remains a major global health burden, often progressing undetected until advanced stages. We applied supervised and unsupervised machine-learning techniques—including random forest classification, k-means clustering, and long short-term memory networks.
Methods: We analyzed the Indian Liver Patient Dataset (583 records) containing age, gender, total and direct bilirubin, alkaline phosphatase, alanine and aspartate aminotransferases, total proteins, albumin, and albumin–globulin ratio. We imputed missing albumin–globulin values with the mean, encoded gender numerically, and created interaction features (age × bilirubin, gender × alkaline phosphatase). We normalized all continuous variables and split the data into 80 percent training and 20 percent testing sets. A random forest classifier distinguished disease-positive from disease-negative cases. To infer severity without predefined labels, we applied k-means clustering (k = 3) on the normalized feature set and mapped each cluster to mild, moderate, or severe stages. We trained a second random forest on these inferred labels to predict severity for new patients. Finally, we generated synthetic five-visit time-series data—spacing visits 30 days apart and adding Gaussian noise—and trained long short-term memory networks to forecast next-visit bilirubin values and severity stages based on the preceding three visits.
Results: The disease classifier reached 81.2 % accuracy on the held-out test set, with precision and recall both above 0.80 for the liver-disease class. Clustering yielded three subgroups of comparable size, each exhibiting distinct biochemical signatures. The severity classifier achieved 99.1 % accuracy in assigning mild, moderate, or severe labels. The bilirubin-forecasting LSTM produced a mean absolute error of 1.07 mg/dL on the test set. The severity-forecasting LSTM achieved 96 % overall accuracy and an F1-score of 0.92 for the severe category. These results demonstrate that combining machine-learning classification, unsupervised clustering, and sequence modeling can accurately detect liver disease, stratify its severity, and anticipate biomarker and stage progression.
Discussion: The results demonstrate the potential of combining classification, clustering, and sequence modeling to support timely diagnosis, personalized risk stratification, and proactive management in liver disease care.

Figure: Figure 1
Disclosures:
Sri Harsha Boppana indicated no relevant financial relationships.
Manaswitha Thota indicated no relevant financial relationships.
Gautam Maddineni indicated no relevant financial relationships.
Sachin Sravan Kumar Komati indicated no relevant financial relationships.
Sarath Chandra Ponnada indicated no relevant financial relationships.
Sai Lakshmi Prasanna Komati indicated no relevant financial relationships.
C. David Mintz indicated no relevant financial relationships.
Sri Harsha Boppana, MBBS, MD1, Manaswitha Thota, MD2, Gautam Maddineni, MD3, Sachin Sravan Kumar Komati, 4, Sarath Chandra Ponnada, 5, Sai Lakshmi Prasanna Komati, MBBS6, C. David Mintz, MD, PhD7. P5867 - A Multi-Stage Machine-Learning Pipeline for Early Detection, Severity Stratification, and Prognostic Forecasting in Liver Disease, ACG 2025 Annual Scientific Meeting Abstracts. Phoenix, AZ: American College of Gastroenterology.
