Tuesday Poster Session
Category: IBD
P5474 - Mapping the Gut-Brain Axis: Genomic, Transcriptomic, and Microbial Overlaps Across IBD, Depression, and Anxiety
Tuesday, October 28, 2025
10:30 AM - 4:00 PM PDT
Location: Exhibit Hall
- ST
Salar Tofighi, MD
Saint Agnes Medical Center
Fresno, CA
Presenting Author(s)
Mahdi Malekpour, MD1, Kiarash Kavari, MD2, Fahimeh Golabi, MD1, Hossein Asadi, MD1, Saranaz Jangjoo, MD1, Farzad Midjani, MD1, Salar Tofighi, MD3
1Shiraz University of Medical Sciences, Shiraz, Fars, Iran; 2shiraz medical university, Shiraz, Fars, Iran; 3Saint Agnes Medical Center, Fresno, CA
Introduction: Inflammatory bowel disease (IBD), depression, and anxiety often co-occur, worsening patient outcomes. Shared mechanisms, including the gut-brain axis, are involved. This study uses large-scale multi-omics data to identify overlapping genomic, transcriptomic, and microbial factors among these disorders.
Methods: GWAS data for IBD, depression, and anxiety were collected from GWASAtlas. Genetic correlations between traits were calculated using Bonferroni correction (p < 0.05). Pleiotropic risk loci were identified via GWASAtlas (−log₁₀(p) > 8). Genes in risk loci were mapped using UCSC Genome Browser with GENCODE v48 and HGNC. Pleiotropic genes found via statistical overlap using GWASAtlas and PheWAS plot (−log₁₀(p) > 8). Trait-associated SNPs were retrieved from the GWAS Catalogue; SNP’s expression effects analyzed using eQTLGen. Microbial-trait links were explored with gutMDisorder, Disbiome, and BGMDB; genetic/immunological links to microbiota explored with GIMICA, GutMGene, and AMADIS. Pathway enrichment of gene sets was conducted using Enrichr-KG, referencing KEGG and GO pathways.
Results: Total of 11 IBD GWAS (191,302 participants), 22 depression (3,146,456), and 4 anxiety (414,570) included. Genetic correlations found between 1 anxiety and 2 UC GWAS, and between 2 IBD and 2 CD with 6 depression GWAS. Risk loci analysis revealed 94 shared loci between IBD and depression (mapping to 2,498 coding/non-coding genes enriched in SLE and neutrophil extracellular trap formation), and 4 loci between IBD and anxiety (27 genes enriched in Leucine response pathways). Statistical overlap showed 37 pleiotropic genes shared by IBD and depression (related to mitotic cytokinesis), and 2 between IBD and anxiety. SNP analysis found 16 shared between IBD and depression, 6 between IBD and anxiety, including 5 common to all three traits. eQTL analysis linked 1,271 SNP–gene expression associations for IBD-depression (enriched in cytokine and interferon signaling) and 69 for IBD-anxiety (linked to autoimmune diseases). Microbiome analysis identified 40 IBD-depression and 27 IBD-anxiety microbial associations, with 18 and 19, respectively, concordant across datasets. These microbial patterns were associated with IBD pathogenesis, and inflammatory and cytokine-related pathways.
Discussion: This large-scale study reveals shared pathways and genetic factors across diseases and omics, while small set of SNVs impacting major pathways that could be used for screenings and drug discovery in further studies.
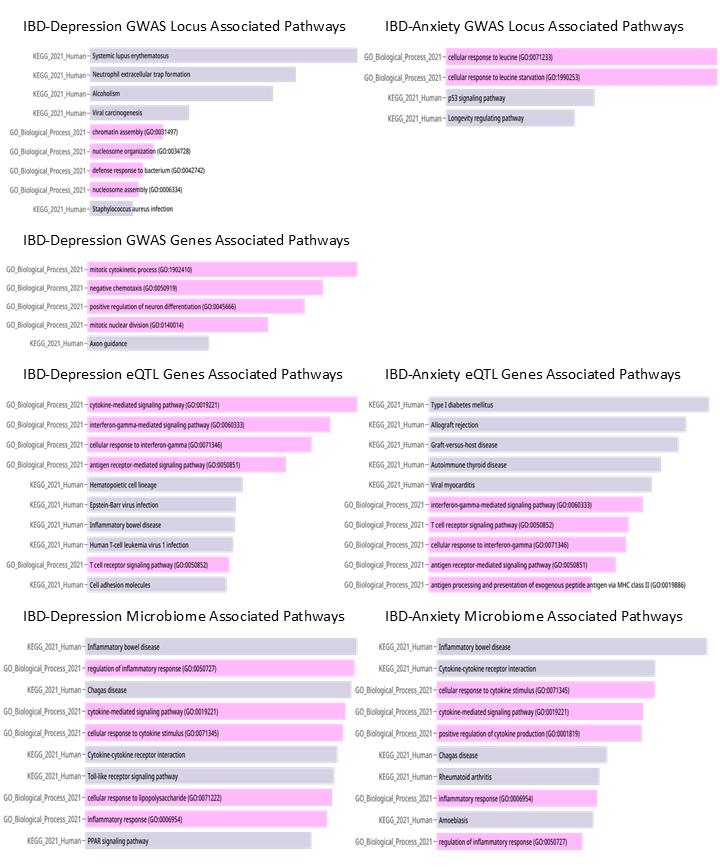
Figure: A. The flow-diagram of study, B. Microbial changes across diseases
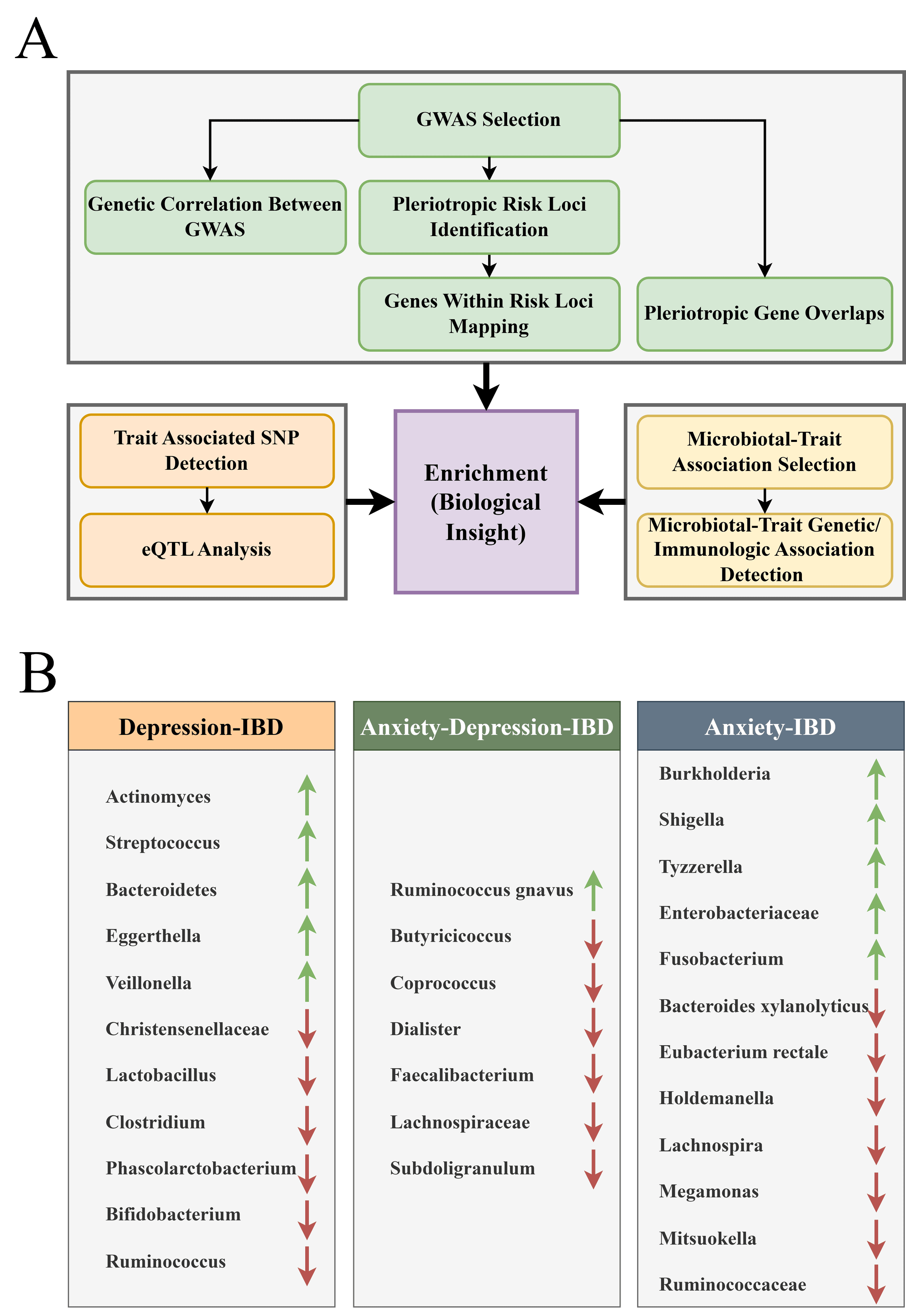
Figure: Enrichment analyses results showing pathways associated with each geneset
Disclosures:
Mahdi Malekpour indicated no relevant financial relationships.
Kiarash Kavari indicated no relevant financial relationships.
Fahimeh Golabi indicated no relevant financial relationships.
Hossein Asadi indicated no relevant financial relationships.
Saranaz Jangjoo indicated no relevant financial relationships.
Farzad Midjani indicated no relevant financial relationships.
Salar Tofighi indicated no relevant financial relationships.
Mahdi Malekpour, MD1, Kiarash Kavari, MD2, Fahimeh Golabi, MD1, Hossein Asadi, MD1, Saranaz Jangjoo, MD1, Farzad Midjani, MD1, Salar Tofighi, MD3. P5474 - Mapping the Gut-Brain Axis: Genomic, Transcriptomic, and Microbial Overlaps Across IBD, Depression, and Anxiety, ACG 2025 Annual Scientific Meeting Abstracts. Phoenix, AZ: American College of Gastroenterology.
1Shiraz University of Medical Sciences, Shiraz, Fars, Iran; 2shiraz medical university, Shiraz, Fars, Iran; 3Saint Agnes Medical Center, Fresno, CA
Introduction: Inflammatory bowel disease (IBD), depression, and anxiety often co-occur, worsening patient outcomes. Shared mechanisms, including the gut-brain axis, are involved. This study uses large-scale multi-omics data to identify overlapping genomic, transcriptomic, and microbial factors among these disorders.
Methods: GWAS data for IBD, depression, and anxiety were collected from GWASAtlas. Genetic correlations between traits were calculated using Bonferroni correction (p < 0.05). Pleiotropic risk loci were identified via GWASAtlas (−log₁₀(p) > 8). Genes in risk loci were mapped using UCSC Genome Browser with GENCODE v48 and HGNC. Pleiotropic genes found via statistical overlap using GWASAtlas and PheWAS plot (−log₁₀(p) > 8). Trait-associated SNPs were retrieved from the GWAS Catalogue; SNP’s expression effects analyzed using eQTLGen. Microbial-trait links were explored with gutMDisorder, Disbiome, and BGMDB; genetic/immunological links to microbiota explored with GIMICA, GutMGene, and AMADIS. Pathway enrichment of gene sets was conducted using Enrichr-KG, referencing KEGG and GO pathways.
Results: Total of 11 IBD GWAS (191,302 participants), 22 depression (3,146,456), and 4 anxiety (414,570) included. Genetic correlations found between 1 anxiety and 2 UC GWAS, and between 2 IBD and 2 CD with 6 depression GWAS. Risk loci analysis revealed 94 shared loci between IBD and depression (mapping to 2,498 coding/non-coding genes enriched in SLE and neutrophil extracellular trap formation), and 4 loci between IBD and anxiety (27 genes enriched in Leucine response pathways). Statistical overlap showed 37 pleiotropic genes shared by IBD and depression (related to mitotic cytokinesis), and 2 between IBD and anxiety. SNP analysis found 16 shared between IBD and depression, 6 between IBD and anxiety, including 5 common to all three traits. eQTL analysis linked 1,271 SNP–gene expression associations for IBD-depression (enriched in cytokine and interferon signaling) and 69 for IBD-anxiety (linked to autoimmune diseases). Microbiome analysis identified 40 IBD-depression and 27 IBD-anxiety microbial associations, with 18 and 19, respectively, concordant across datasets. These microbial patterns were associated with IBD pathogenesis, and inflammatory and cytokine-related pathways.
Discussion: This large-scale study reveals shared pathways and genetic factors across diseases and omics, while small set of SNVs impacting major pathways that could be used for screenings and drug discovery in further studies.

Figure: A. The flow-diagram of study, B. Microbial changes across diseases

Figure: Enrichment analyses results showing pathways associated with each geneset
Disclosures:
Mahdi Malekpour indicated no relevant financial relationships.
Kiarash Kavari indicated no relevant financial relationships.
Fahimeh Golabi indicated no relevant financial relationships.
Hossein Asadi indicated no relevant financial relationships.
Saranaz Jangjoo indicated no relevant financial relationships.
Farzad Midjani indicated no relevant financial relationships.
Salar Tofighi indicated no relevant financial relationships.
Mahdi Malekpour, MD1, Kiarash Kavari, MD2, Fahimeh Golabi, MD1, Hossein Asadi, MD1, Saranaz Jangjoo, MD1, Farzad Midjani, MD1, Salar Tofighi, MD3. P5474 - Mapping the Gut-Brain Axis: Genomic, Transcriptomic, and Microbial Overlaps Across IBD, Depression, and Anxiety, ACG 2025 Annual Scientific Meeting Abstracts. Phoenix, AZ: American College of Gastroenterology.
