Tuesday Poster Session
Category: General Endoscopy
P5142 - Privacy-Preserving Approach to Endoscopic Report Data Extraction Using General-Purpose Versus Medically Fine-Tuned Locally Hosted Large Language Models
Tuesday, October 28, 2025
10:30 AM - 4:00 PM PDT
Location: Exhibit Hall

Rebecca Fine, MD
University of Kentucky College of Medicine
Lexington, KY
Presenting Author(s)
Rebecca Fine, MD1, Hannah Darnell, DO1, Ismail S. Bahaaeldeen, MD2
1University of Kentucky College of Medicine, Lexington, KY; 2University of Kentucky, Lexington, KY
Introduction: Endoscopy data extraction is essential for research, clinical care, and quality assurance, yet remains challenging due to the partially unstructured nature and varying formats of endoscopy reports across different software platforms. Large language models (LLMs) have recently been leveraged for this task, with the expectation that models fine-tuned on medical data should outperform general-purpose versions. However, formal comparisons of these approaches are lacking.
Methods: This study compares the performance, efficiency, and environmental impact of two general-purpose LLMs (Gemma3:4b and Llama3:8b) and their medically fine-tuned counterparts (Med-Gemma and OpenBio LLM). All models were executed locally on a consumer-grade desktop (Intel i5-10500 CPU,16 GB RAM, Windows 10, no GPU), ensuring patient data privacy by avoiding external data transfer. We evaluated 88 endoscopy reports for esophageal food impaction using Epic Lumens' EndoManager® Report Writer, submitting each report to the models along with detailed instructions for 23 variables via a programmatic interface. We calculated the percentage agreement of each model with human extraction ground truth, and we used CodeCarbon Python package (2.7.4) to estimate energy consumption.
Results: Correct extraction rates ranged from 81% to 87.2% (Figure 1). The general-purpose Gemma3 model achieved the highest accuracy (87.2%), significantly outperforming its medically fine-tuned version (83.4%, p=0.004). Similarly, general-purpose Llama3 (86.3%) performed better than its fine-tuned counterpart (81%, p=0.01). Across all models, the calculated extraction accuracy for free-text variable extraction was lower than Boolean variable extraction (75.5% versus 87.6%). The two Gemma models had slightly faster processing compared to the Llama3 models, all p < 0.01 (Figure 2). Energy consumption and CO₂ emissions were significantly lower for the Gemma models compared to Llama3, however, with no significant difference between the two Gemma variants.
Discussion: Our approach achieves modest extraction accuracy with full data privacy via 100% local processing, making it a viable option for endoscopy data extraction. Unexpectedly, general-purpose models outperformed fine-tuned domain-specific ones, prompting a reconsideration of model selection strategies. The smaller 4B Gemma model also outperformed the 8B variant in accuracy, speed, and efficiency, highlighting the promise of lightweight models for effective and sustainable clinical data processing.
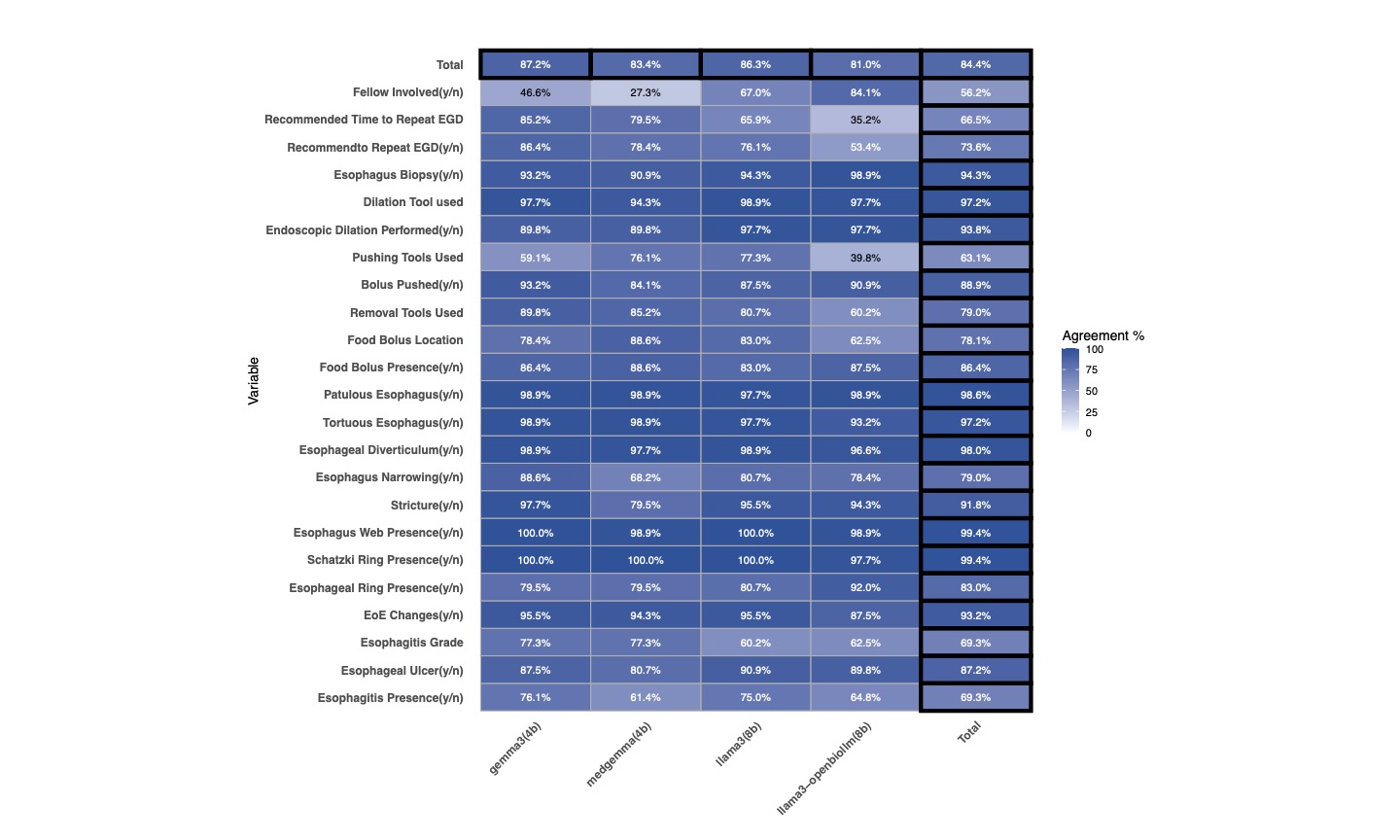
Figure: Figure 1: A heatmap depicting the variable agreement for each model.
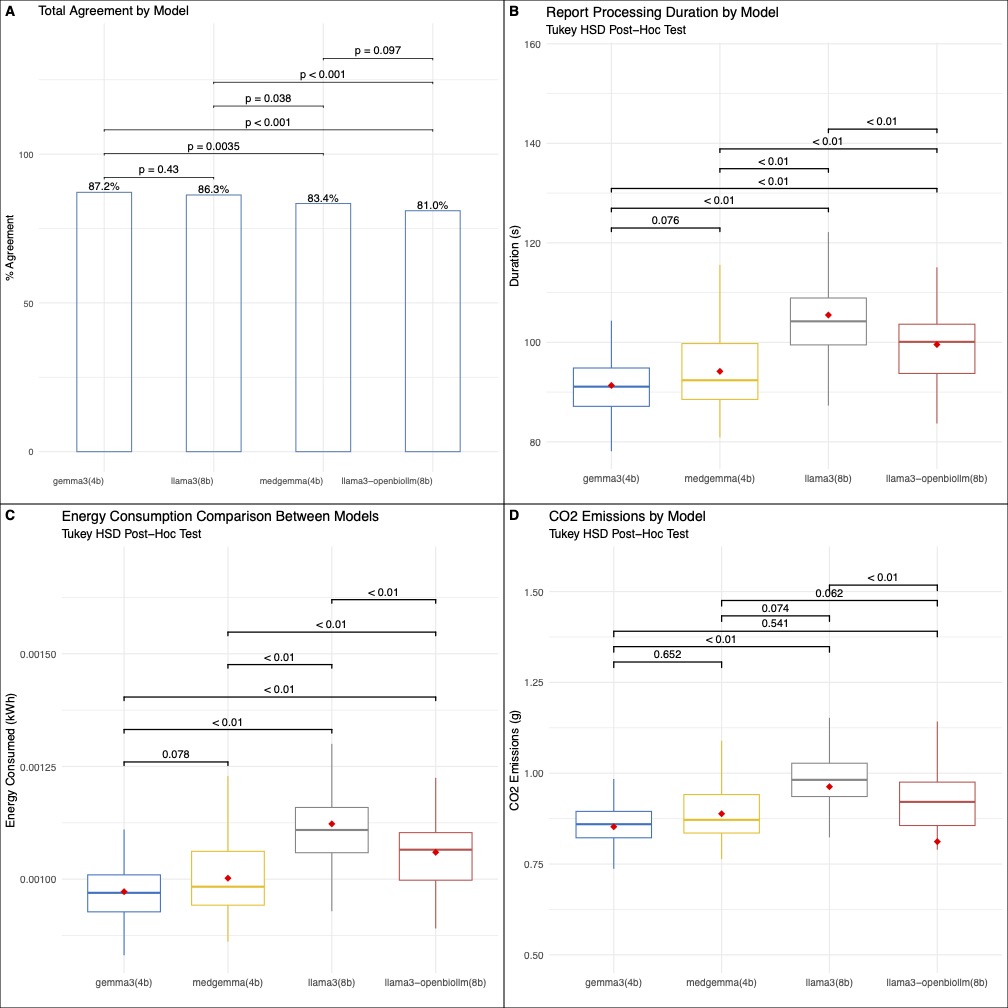
Figure: Figure 2: A depiction of variable agreement, processing duration, energy consumption, and CO2 emissions for each model.
Disclosures:
Rebecca Fine indicated no relevant financial relationships.
Hannah Darnell indicated no relevant financial relationships.
Ismail Bahaaeldeen indicated no relevant financial relationships.
Rebecca Fine, MD1, Hannah Darnell, DO1, Ismail S. Bahaaeldeen, MD2. P5142 - Privacy-Preserving Approach to Endoscopic Report Data Extraction Using General-Purpose Versus Medically Fine-Tuned Locally Hosted Large Language Models, ACG 2025 Annual Scientific Meeting Abstracts. Phoenix, AZ: American College of Gastroenterology.
1University of Kentucky College of Medicine, Lexington, KY; 2University of Kentucky, Lexington, KY
Introduction: Endoscopy data extraction is essential for research, clinical care, and quality assurance, yet remains challenging due to the partially unstructured nature and varying formats of endoscopy reports across different software platforms. Large language models (LLMs) have recently been leveraged for this task, with the expectation that models fine-tuned on medical data should outperform general-purpose versions. However, formal comparisons of these approaches are lacking.
Methods: This study compares the performance, efficiency, and environmental impact of two general-purpose LLMs (Gemma3:4b and Llama3:8b) and their medically fine-tuned counterparts (Med-Gemma and OpenBio LLM). All models were executed locally on a consumer-grade desktop (Intel i5-10500 CPU,16 GB RAM, Windows 10, no GPU), ensuring patient data privacy by avoiding external data transfer. We evaluated 88 endoscopy reports for esophageal food impaction using Epic Lumens' EndoManager® Report Writer, submitting each report to the models along with detailed instructions for 23 variables via a programmatic interface. We calculated the percentage agreement of each model with human extraction ground truth, and we used CodeCarbon Python package (2.7.4) to estimate energy consumption.
Results: Correct extraction rates ranged from 81% to 87.2% (Figure 1). The general-purpose Gemma3 model achieved the highest accuracy (87.2%), significantly outperforming its medically fine-tuned version (83.4%, p=0.004). Similarly, general-purpose Llama3 (86.3%) performed better than its fine-tuned counterpart (81%, p=0.01). Across all models, the calculated extraction accuracy for free-text variable extraction was lower than Boolean variable extraction (75.5% versus 87.6%). The two Gemma models had slightly faster processing compared to the Llama3 models, all p < 0.01 (Figure 2). Energy consumption and CO₂ emissions were significantly lower for the Gemma models compared to Llama3, however, with no significant difference between the two Gemma variants.
Discussion: Our approach achieves modest extraction accuracy with full data privacy via 100% local processing, making it a viable option for endoscopy data extraction. Unexpectedly, general-purpose models outperformed fine-tuned domain-specific ones, prompting a reconsideration of model selection strategies. The smaller 4B Gemma model also outperformed the 8B variant in accuracy, speed, and efficiency, highlighting the promise of lightweight models for effective and sustainable clinical data processing.

Figure: Figure 1: A heatmap depicting the variable agreement for each model.

Figure: Figure 2: A depiction of variable agreement, processing duration, energy consumption, and CO2 emissions for each model.
Disclosures:
Rebecca Fine indicated no relevant financial relationships.
Hannah Darnell indicated no relevant financial relationships.
Ismail Bahaaeldeen indicated no relevant financial relationships.
Rebecca Fine, MD1, Hannah Darnell, DO1, Ismail S. Bahaaeldeen, MD2. P5142 - Privacy-Preserving Approach to Endoscopic Report Data Extraction Using General-Purpose Versus Medically Fine-Tuned Locally Hosted Large Language Models, ACG 2025 Annual Scientific Meeting Abstracts. Phoenix, AZ: American College of Gastroenterology.

