Monday Poster Session
Category: Colon
P2435 - Genomic Profiling-Based Prognosis and Survival of Colorectal Carcinoma: A Mutation and Survival Analysis from CBioPortal

Urvish Patel, MD, MPH (he/him/his)
Icahn School of Medicine at Mount Sinai
New York, NY
Presenting Author(s)
1Icahn School of Medicine at Mount Sinai, New York, NY; 2GMERS Medical College and Hospital, Junagadh, Junagadh, Gujarat, India; 3Infirmary Health, Mobile, AL; 4GMERS Medical College Gotri, Vadodara, Gujarat, India; 5Harlem Hospital Center, New York, NY; 6Cleveland Clinic Akron General Hospital, Akron, OH; 7Medical College Baroda, Vadodara, Gujarat, Vadodara, Gujarat, India; 8Avalon University School of Medicine, Youngstown, OH; 9Western Reserve Hospital, Cuyahoga Falls, OH; 10Mercy Health St Vincent Medical Center, Toledo, OH; 11Memorial Hospital at Gulfport Internal Medicine Residency, Gulfport, MS; 12University of Texas MD Anderson Cancer Center, Houston, TX
Introduction:
The colorectal carcinoma (CRC) is the most prevalent cancer, and carries a poor prognosis. Information regarding whole genome sequencing, a prognostic prediction system, and overall survival in CRC is limited. While mutation of TP53 and APC are commonly implicated in CRC, EGFR, MLH1, MSH2, MSH6, PMS2, TGFBR2, KRAS, SMAD4, PTEN, and BRAF genes have limited literature on their correlation to CRC. Hence, we aimed to analyze the genomic profile, epidemiological characteristics, and overall survival of CRC.
Methods:
We utilized cBioPortal cancer genomics [The Cancer Genome Atlas (TCGA) PanCancer Atlas, TCGA Firehose Legacy, and TCGA Nature 2012, and MSKCC] to study the genetic mutations associated with CRC. Epidemiological characteristics and genetic profiles were evaluated, and a query was generated to calculate the 10-year survival rate with the most common mutations. Log-rank test and Kaplan–Meier estimator were used in analyzing the survival function. Patients with two or more overlapping mutations (435) were excluded. Our study found that the MLH1 gene was associated with the worst prognosis in CRC with a median survival of 38 months. This study may provide insight into targeted therapies for CRC and improve the prognosis of these patients.
Results: We identified 5,272 samples (5,062 patients) with CRC. Out of which 4,628 (87.8%) had one or more mutations. The most common mutations were APC (72.5%), TP53 (67.1%), TTN (44.8%), KRAS (42.5%), SYNE1 (25.2%), MUC16 (23.7%), FAT4 (19.8%), PIK3CA (19.7%), FLG (19.1%), LRP1B (17.0%). The most common treatment utilized for CRC was Fluorouracil, Leucovorin, Oxaliplatin, Bevacizumab, and Radiation. 1951 (37% samples) patients were between the ages of 50-80 years. 2442 (46.3%) had primary CRC and 14.1% had metastasis. 50.7% were males, 23.3% were Caucasian, and 2.5% were African American. Median survival of patients with APC (49 months), TP53 (39 months), MLH1 (38 months), MSH6 (22 months), SMAD4 (49 months), BRAF (31 months), EPCAM (41 months), and BMPR1A (34 months) mutations was lower in comparison with the unaltered mutation group (62 months) (p< 0.0001). (Table 1) In survival analysis, MLH1 was associated with the lowest survival after initial diagnosis at 120 months (Figure 1). (p=2.98e-3)
Discussion: 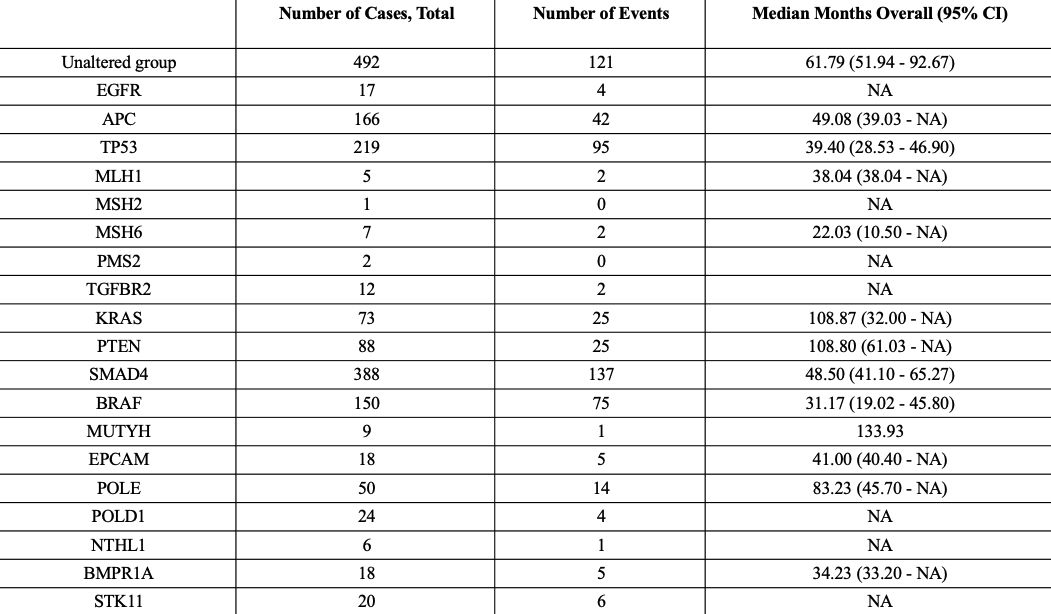
Figure: Log-rank test and Kaplan–Meier estimator for analyzing the survival associated with genetic mutation 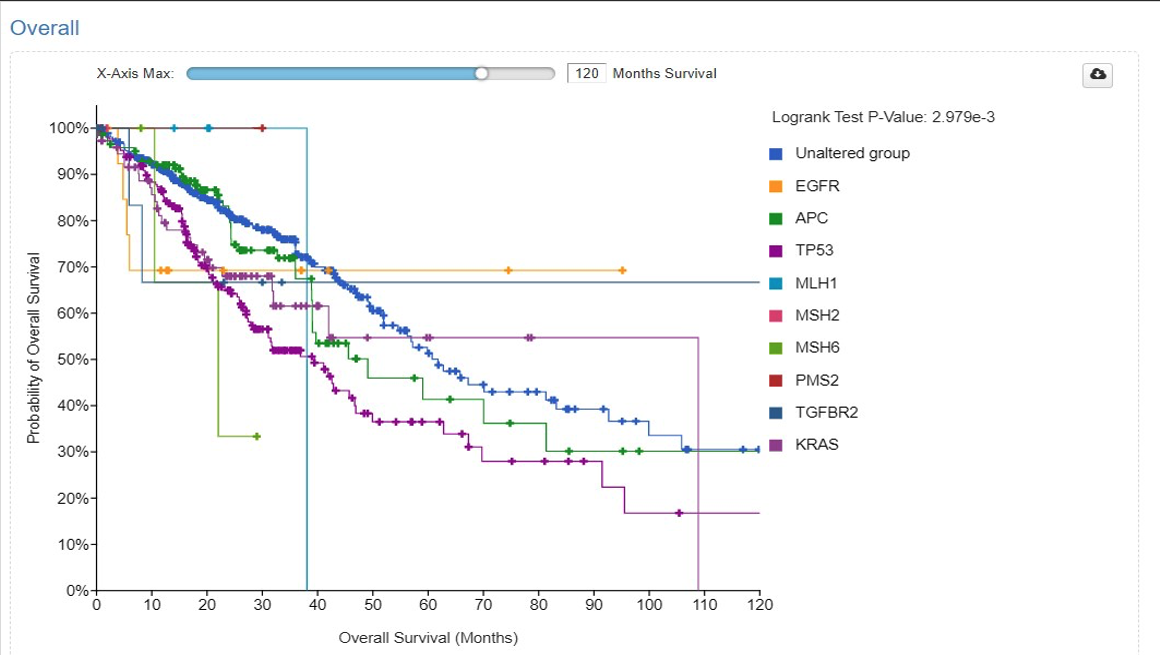
Figure: Median Overall Survival in Colorectal Cancer Patients Based on Specific Gene Mutations
Disclosures:
Urvish Patel indicated no relevant financial relationships.
Smit Kotadiya indicated no relevant financial relationships.
Meghana Kakarla indicated no relevant financial relationships.
Anshi Gelani indicated no relevant financial relationships.
Dhruva Chauhan indicated no relevant financial relationships.
Vyshnavi Iswaravaka indicated no relevant financial relationships.
Namra Gohil indicated no relevant financial relationships.
Jay Prashar indicated no relevant financial relationships.
Zarna Bambhroliya indicated no relevant financial relationships.
Vijay Kumar Doddapaneni indicated no relevant financial relationships.
Ansh Purohit indicated no relevant financial relationships.
Ayush Gandhi indicated no relevant financial relationships.
Urvish Patel, MD, MPH1, Smit Kotadiya, MBBS2, Meghana Kakarla, MD3, Anshi M. Gelani, MBBS4, Dhruva Chauhan, MD5, Vyshnavi Iswaravaka, MD6, Namra Gohil, MBBS7, Jay Prashar, MD8, Zarna Bambhroliya, MD9, Vijay Kumar Doddapaneni, MD10, Ansh Purohit, MD11, Ayush Gandhi, MD12. P2435 - Genomic Profiling-Based Prognosis and Survival of Colorectal Carcinoma: A Mutation and Survival Analysis from CBioPortal, ACG 2025 Annual Scientific Meeting Abstracts. Phoenix, AZ: American College of Gastroenterology.
