Monday Poster Session
Category: IBD
P3212 - Ulcerative Colitis Advanced Therapy Timeline Prediction Using Large Language Models
Monday, October 27, 2025
10:30 AM - 4:00 PM PDT
Location: Exhibit Hall

Yuntao Zou, MD
University of California San Francisco
San Francisco, CA
Presenting Author(s)
Yuntao Zou, MD, Mao-Yuan Chen, MD, Richard Yim, MS, Peter Washington, PhD, Yulin Hswen, ScD, Vivek Rudrapatna, MD, PhD
University of California San Francisco, San Francisco, CA
Introduction: Accurately reconstructing medication timelines is essential across chronic diseases such as ulcerative colitis (UC), but manual chart review is laborious and structured data alone often misclassifies key dates. The start date is pivotal—it defines therapy sequencing and subsequent milestones (e.g., end of induction, one-year follow-up). We therefore developed a large language model (LLM)-based method that combines unstructured data and structured data to precisely predict each advanced therapy’s start date.
Methods: We identified 193 UC patients seen at University of California, San Francisco (UCSF) Inflammatory Bowel Disease Center (2018–2023) and split them into training (n=97) and test (n=96) sets. Our goal was to predict the start dates of advanced therapies initiated at UCSF using structured inputs (orders, labs, administrations, procedures) and unstructured clinical notes processed by a HIPAA-compliant GPT-4o. Because our database only captures UCSF encounters, we first classified whether a medication was started at UCSF, then estimated its start date. Four models were compared against manual annotation:
1. Structured data via mixed-effects model
2. Unstructured data processed by GPT-4o only
3. Mixed-effects model combining structured data with GPT-4o–processed notes
4. Combined structured and unstructured data processed by GPT-4o only
Results: Model 4 most accurately identified UCSF-initiated medications (sensitivity 94.4%, specificity 90.1%), outperforming models 1–3 (sens./spec.: 74.2%/71.6%, 79.8%/76.2%, 86.4%/81.2%). Among correctly classified UCSF starts, Model 4 also had the lowest mean absolute start-date error (30.3 days vs. 119.1, 74.3, 73.5 days for models 1–3; p < 0.01). Models 2 and 3 both outperformed model 1 (p < 0.01) but did not differ from each other (p = 0.94). Using a one-month cutoff, Model 4 was accurate 90.9% of the time (vs. 43.2%, 65.9%, 79.6%), and using a two-week cutoff, Model 4 remained best at 81.8% (vs. 38.6%, 68.2%, 61.4%).
Discussion: By processing structured data and unstructured clinical notes together only with an LLM, Model 4 minimized start-date prediction error more effectively than models using only structured data, only unstructured data, or a mixed-effects approach with both. It achieved over 90% accuracy within one month and over 80% within two weeks. While demonstrated in UC, this scalable method applies broadly to medication-use studies across diseases.
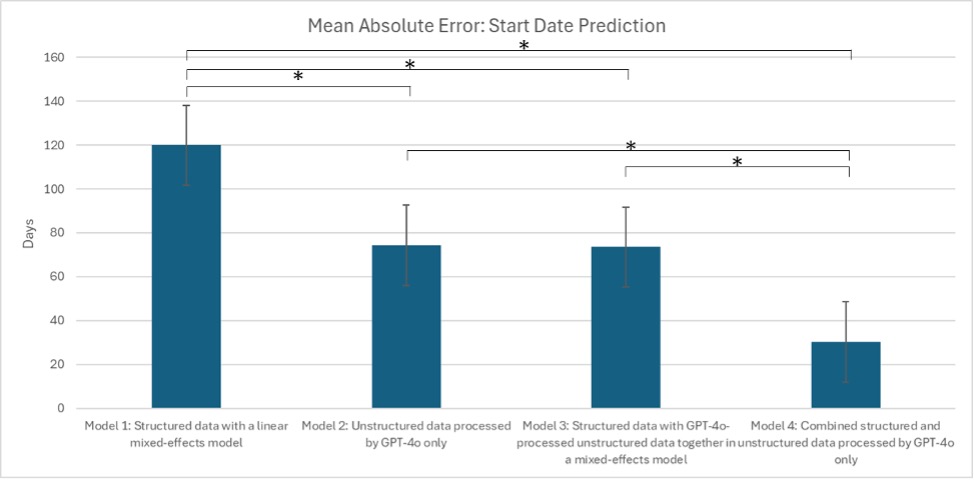
Figure: Figure 1. Mean Absolute Error: Start Date Prediction
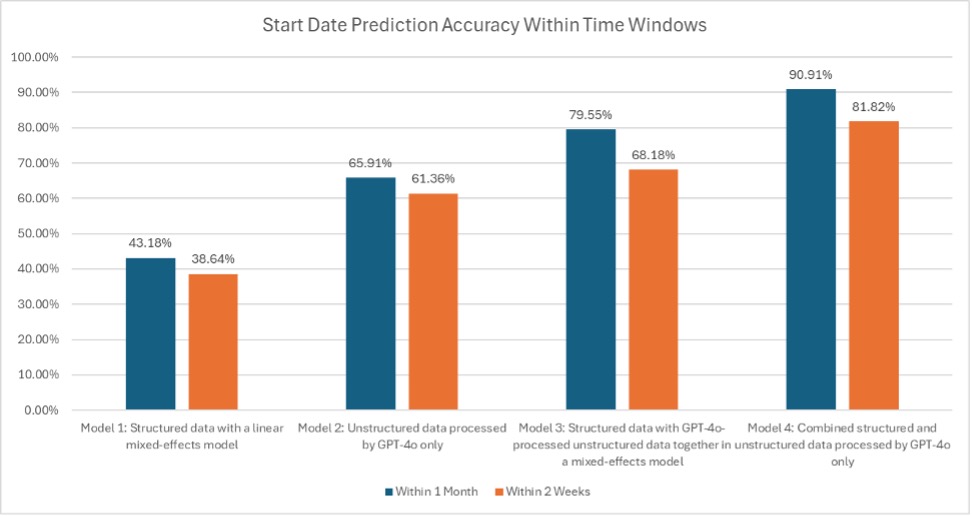
Figure: Figure 2. Start Date Prediction Accuracy Within Time Windows
Disclosures:
Yuntao Zou indicated no relevant financial relationships.
Mao-Yuan Chen indicated no relevant financial relationships.
Richard Yim indicated no relevant financial relationships.
Peter Washington indicated no relevant financial relationships.
Yulin Hswen indicated no relevant financial relationships.
Vivek Rudrapatna: Acucare – Advisory Committee/Board Member. Blueprint Medicines – Grant/Research Support. Data Unite – Advisory Committee/Board Member. Genentech – Grant/Research Support. Ironwood – Payment or honoraria for lectures, presentations, speakers bureaus, manuscript writing or educational events. Merck – Grant/Research Support. Microsoft – Grant/Research Support. Mitsubishi Tanabe – Grant/Research Support. Natera – Payment or honoraria for lectures, presentations, speakers bureaus, manuscript writing or educational events. Stryker – Grant/Research Support. Takeda – Grant/Research Support. ZebraMD – Advisory Committee/Board Member.
Yuntao Zou, MD, Mao-Yuan Chen, MD, Richard Yim, MS, Peter Washington, PhD, Yulin Hswen, ScD, Vivek Rudrapatna, MD, PhD. P3212 - Ulcerative Colitis Advanced Therapy Timeline Prediction Using Large Language Models, ACG 2025 Annual Scientific Meeting Abstracts. Phoenix, AZ: American College of Gastroenterology.
University of California San Francisco, San Francisco, CA
Introduction: Accurately reconstructing medication timelines is essential across chronic diseases such as ulcerative colitis (UC), but manual chart review is laborious and structured data alone often misclassifies key dates. The start date is pivotal—it defines therapy sequencing and subsequent milestones (e.g., end of induction, one-year follow-up). We therefore developed a large language model (LLM)-based method that combines unstructured data and structured data to precisely predict each advanced therapy’s start date.
Methods: We identified 193 UC patients seen at University of California, San Francisco (UCSF) Inflammatory Bowel Disease Center (2018–2023) and split them into training (n=97) and test (n=96) sets. Our goal was to predict the start dates of advanced therapies initiated at UCSF using structured inputs (orders, labs, administrations, procedures) and unstructured clinical notes processed by a HIPAA-compliant GPT-4o. Because our database only captures UCSF encounters, we first classified whether a medication was started at UCSF, then estimated its start date. Four models were compared against manual annotation:
1. Structured data via mixed-effects model
2. Unstructured data processed by GPT-4o only
3. Mixed-effects model combining structured data with GPT-4o–processed notes
4. Combined structured and unstructured data processed by GPT-4o only
Results: Model 4 most accurately identified UCSF-initiated medications (sensitivity 94.4%, specificity 90.1%), outperforming models 1–3 (sens./spec.: 74.2%/71.6%, 79.8%/76.2%, 86.4%/81.2%). Among correctly classified UCSF starts, Model 4 also had the lowest mean absolute start-date error (30.3 days vs. 119.1, 74.3, 73.5 days for models 1–3; p < 0.01). Models 2 and 3 both outperformed model 1 (p < 0.01) but did not differ from each other (p = 0.94). Using a one-month cutoff, Model 4 was accurate 90.9% of the time (vs. 43.2%, 65.9%, 79.6%), and using a two-week cutoff, Model 4 remained best at 81.8% (vs. 38.6%, 68.2%, 61.4%).
Discussion: By processing structured data and unstructured clinical notes together only with an LLM, Model 4 minimized start-date prediction error more effectively than models using only structured data, only unstructured data, or a mixed-effects approach with both. It achieved over 90% accuracy within one month and over 80% within two weeks. While demonstrated in UC, this scalable method applies broadly to medication-use studies across diseases.

Figure: Figure 1. Mean Absolute Error: Start Date Prediction

Figure: Figure 2. Start Date Prediction Accuracy Within Time Windows
Disclosures:
Yuntao Zou indicated no relevant financial relationships.
Mao-Yuan Chen indicated no relevant financial relationships.
Richard Yim indicated no relevant financial relationships.
Peter Washington indicated no relevant financial relationships.
Yulin Hswen indicated no relevant financial relationships.
Vivek Rudrapatna: Acucare – Advisory Committee/Board Member. Blueprint Medicines – Grant/Research Support. Data Unite – Advisory Committee/Board Member. Genentech – Grant/Research Support. Ironwood – Payment or honoraria for lectures, presentations, speakers bureaus, manuscript writing or educational events. Merck – Grant/Research Support. Microsoft – Grant/Research Support. Mitsubishi Tanabe – Grant/Research Support. Natera – Payment or honoraria for lectures, presentations, speakers bureaus, manuscript writing or educational events. Stryker – Grant/Research Support. Takeda – Grant/Research Support. ZebraMD – Advisory Committee/Board Member.
Yuntao Zou, MD, Mao-Yuan Chen, MD, Richard Yim, MS, Peter Washington, PhD, Yulin Hswen, ScD, Vivek Rudrapatna, MD, PhD. P3212 - Ulcerative Colitis Advanced Therapy Timeline Prediction Using Large Language Models, ACG 2025 Annual Scientific Meeting Abstracts. Phoenix, AZ: American College of Gastroenterology.
