Monday Poster Session
Category: Small Intestine
P4027 - Deep Learning Algorithm for Duodenal Biopsy Classification
Monday, October 27, 2025
10:30 AM - 4:00 PM PDT
Location: Exhibit Hall

Claire Jansson-Knodell, MD
Cleveland Clinic Foundation
Cleveland, OH
Presenting Author(s)
Award: ACG Presidential Poster Award
Claire Jansson-Knodell, MD, Erica Savage, MD, Katherine Poissant, BS, Ivan Yu, , Matthew Kerosky, , Rich Fong, MBA, Scott Robertson, MD, Alberto Rubio Tapia, MD
Cleveland Clinic Foundation, Cleveland, OH
Introduction: Duodenal biopsy combined with tissue transglutaminase (TTG-IgA) antibodies is the gold standard for celiac disease diagnosis. Issues with biopsy include artifacts, poorly oriented specimens, and inter-observer variability. Artificial intelligence has the potential to address these challenges. We aimed to determine if deep learning, image analysis using artificial intelligence, can be used on duodenal biopsies to accurately diagnose celiac disease.
Methods: The target study population were individuals with a duodenal biopsy and TTG-IgA antibody test. Inclusion criteria were adults ≥18 years. Exclusion criteria were those following a gluten-free diet. The study period was 2008-2021. Patients were grouped into 5 categories: controls, celiac disease, increased intraepithelial lymphocytes (IELs) only, IELs with a positive TTG-IgA antibody, and seronegative villous atrophy. Biopsy were stained with hematoxylin and eosin. Whole slide images were obtained and placed on a High-Performance Computing node with a graphics processing unit. Clustering-constrained attention multiple-instance learning was the deep learning algorithm used. A patch size of 256x256 pixels was used for the segmentation step. The dataset was split into training, validation, and testing splits with 10-fold cross validation. Accuracy and area under the receiver operating characteristics curve (AUC) were reported for each fold.
Results: A total of 2,392 patients’ whole slide images were included: 1290 controls, 414 celiac disease, 441 IELs only, 84 IELs with a positive TTG-IgA, and 163 with seronegative villous atrophy. For all categories, the average AUC was 0.849 with an accuracy of 71%. The best performing fold had an AUC of 0.884 with 75% accuracy. The confusion matrix for this fold (Figure 1) demonstrates difficulty differentiating between IELs with a positive TTG-IgA antibody and other classifications. The model reliably differentiated celiac disease from controls average with AUC 0.992 and accuracy 97%. To distinguish celiac disease from seronegative villous atrophy the average AUC was 0.810 with accuracy 76%.
Discussion: This deep learning model can discriminate between celiac disease and normal histology. It can also separate patients into 5 classifications, representing a promising tool that would benefit from further optimization to improve diagnostic accuracy, reduce the manual pathology review burden, and enhance resource management.
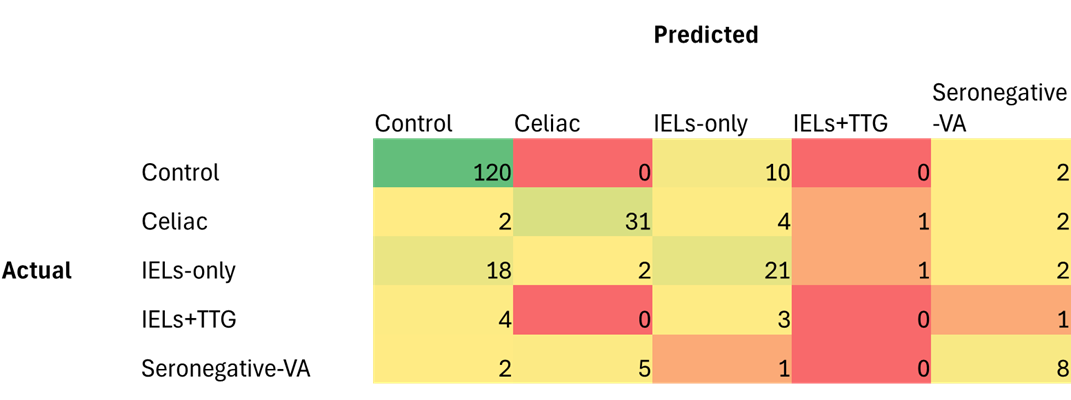
Figure: Figure 1. Confusion Matrix for Best Performing Fold Overall
Disclosures:
Claire Jansson-Knodell: AGA-Takeda Pharmaceuticals – Grant/Research Support. Exact Sciences – Stock-publicly held company(excluding mutual/index funds). Johnson&Johnson – Stock-publicly held company(excluding mutual/index funds). Medtronic – Stock-publicly held company(excluding mutual/index funds). United Health Group – Stock-publicly held company(excluding mutual/index funds).
Erica Savage indicated no relevant financial relationships.
Katherine Poissant indicated no relevant financial relationships.
Ivan Yu indicated no relevant financial relationships.
Matthew Kerosky indicated no relevant financial relationships.
Rich Fong indicated no relevant financial relationships.
Scott Robertson indicated no relevant financial relationships.
Alberto Rubio Tapia indicated no relevant financial relationships.
Claire Jansson-Knodell, MD, Erica Savage, MD, Katherine Poissant, BS, Ivan Yu, , Matthew Kerosky, , Rich Fong, MBA, Scott Robertson, MD, Alberto Rubio Tapia, MD. P4027 - Deep Learning Algorithm for Duodenal Biopsy Classification, ACG 2025 Annual Scientific Meeting Abstracts. Phoenix, AZ: American College of Gastroenterology.
Claire Jansson-Knodell, MD, Erica Savage, MD, Katherine Poissant, BS, Ivan Yu, , Matthew Kerosky, , Rich Fong, MBA, Scott Robertson, MD, Alberto Rubio Tapia, MD
Cleveland Clinic Foundation, Cleveland, OH
Introduction: Duodenal biopsy combined with tissue transglutaminase (TTG-IgA) antibodies is the gold standard for celiac disease diagnosis. Issues with biopsy include artifacts, poorly oriented specimens, and inter-observer variability. Artificial intelligence has the potential to address these challenges. We aimed to determine if deep learning, image analysis using artificial intelligence, can be used on duodenal biopsies to accurately diagnose celiac disease.
Methods: The target study population were individuals with a duodenal biopsy and TTG-IgA antibody test. Inclusion criteria were adults ≥18 years. Exclusion criteria were those following a gluten-free diet. The study period was 2008-2021. Patients were grouped into 5 categories: controls, celiac disease, increased intraepithelial lymphocytes (IELs) only, IELs with a positive TTG-IgA antibody, and seronegative villous atrophy. Biopsy were stained with hematoxylin and eosin. Whole slide images were obtained and placed on a High-Performance Computing node with a graphics processing unit. Clustering-constrained attention multiple-instance learning was the deep learning algorithm used. A patch size of 256x256 pixels was used for the segmentation step. The dataset was split into training, validation, and testing splits with 10-fold cross validation. Accuracy and area under the receiver operating characteristics curve (AUC) were reported for each fold.
Results: A total of 2,392 patients’ whole slide images were included: 1290 controls, 414 celiac disease, 441 IELs only, 84 IELs with a positive TTG-IgA, and 163 with seronegative villous atrophy. For all categories, the average AUC was 0.849 with an accuracy of 71%. The best performing fold had an AUC of 0.884 with 75% accuracy. The confusion matrix for this fold (Figure 1) demonstrates difficulty differentiating between IELs with a positive TTG-IgA antibody and other classifications. The model reliably differentiated celiac disease from controls average with AUC 0.992 and accuracy 97%. To distinguish celiac disease from seronegative villous atrophy the average AUC was 0.810 with accuracy 76%.
Discussion: This deep learning model can discriminate between celiac disease and normal histology. It can also separate patients into 5 classifications, representing a promising tool that would benefit from further optimization to improve diagnostic accuracy, reduce the manual pathology review burden, and enhance resource management.

Figure: Figure 1. Confusion Matrix for Best Performing Fold Overall
Disclosures:
Claire Jansson-Knodell: AGA-Takeda Pharmaceuticals – Grant/Research Support. Exact Sciences – Stock-publicly held company(excluding mutual/index funds). Johnson&Johnson – Stock-publicly held company(excluding mutual/index funds). Medtronic – Stock-publicly held company(excluding mutual/index funds). United Health Group – Stock-publicly held company(excluding mutual/index funds).
Erica Savage indicated no relevant financial relationships.
Katherine Poissant indicated no relevant financial relationships.
Ivan Yu indicated no relevant financial relationships.
Matthew Kerosky indicated no relevant financial relationships.
Rich Fong indicated no relevant financial relationships.
Scott Robertson indicated no relevant financial relationships.
Alberto Rubio Tapia indicated no relevant financial relationships.
Claire Jansson-Knodell, MD, Erica Savage, MD, Katherine Poissant, BS, Ivan Yu, , Matthew Kerosky, , Rich Fong, MBA, Scott Robertson, MD, Alberto Rubio Tapia, MD. P4027 - Deep Learning Algorithm for Duodenal Biopsy Classification, ACG 2025 Annual Scientific Meeting Abstracts. Phoenix, AZ: American College of Gastroenterology.

