Monday Poster Session
Category: Esophagus
P2739 - Large Language Models Combined With a Single Diagnostic Code Detect Eosinophilic Esophagitis in Electronic Health Records With High Accuracy
Monday, October 27, 2025
10:30 AM - 4:00 PM PDT
Location: Exhibit Hall

Corey J. Ketchem, MD
University of Pennsylvania Health System
CHICAGO, IL
Presenting Author(s)
Corey J. Ketchem, MD1, Uğurcan Vurgun, PhD, MA2, Agnes Wang, MS3, Sunil Thomas, MBA4, Ashley Batugo, BS4, Gary Falk, MD, MS5, Kristle L.. Lynch, MD6, Evan S. Dellon, MD, MPH7, Danielle Mowery, PhD, MS8, James Lewis, MD, MSCE9
1University of Pennsylvania Health System, Philadelphia, PA; 2University of Pennsylvania, Mill Creek, WA; 3Univeristy of Pennsylvania, Philadelphia, PA; 4University of Pennsylvania, Philadelphia, PA; 5Division of Gastroenterology and Hepatology, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA; 6The University of Pennsylvania, Philadelphia, PA; 7Center for Esophageal Diseases and Swallowing, University of North Carolina School of Medicine, Chapel Hill, NC; 8Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA; 9University of Penn, Philadelphia, PA
Introduction: Large-scale epidemiologic research for eosinophilic esophagitis (EoE) is hampered by variable accuracy of International Classification of Diseases (ICD) codes and case identification algorithms. We aimed to develop and validate a natural language processing (NLP) pipeline using large language models (LLMs) to identify EoE features and diagnosis from unstructured text, comparing performance with ICD codes and ICD+LLM combination.
Methods: We identified gastrointestinal pathology reports with any mention of “eosinophil” paired with preceding GI clinic notes (Figure 1A). 300 randomly selected patients were divided into training (n=200, 56 with EoE) and testing (n=100, 36 with EoE) sets. Manual chart review was used to assign an EoE reference standard. LLM prompt development used a human-in-the-loop approach with iterative refinements (Figure 1B). Training concluded once the LLM-assigned diagnosis exceeded the F1 score (harmonic mean of precision and recall) of ICD codes. Using the test set, we compared performance of ICD codes, LLM-derived diagnostic features, LLM-assigned diagnosis, and a combined LLM+ICD. Performance metrics included sensitivity (recall), PPV (precision), and F1 score. Nonparametric bootstrap resampling (1,000 replicates) was used to estimate 95% confidence intervals with statistical significance based on whether they excluded zero.
Results: In the training set, LLM-derived diagnostic features demonstrated the highest sensitivity (0.98, 95% CI: 0.95,1.00), while LLM-assigned diagnosis had the highest PPV (0.92, 95% CI: 0.84-1.0) and specificity (0.97, 95% CI: 0.95,1.00]) (Table 1). Comparably high F1 scores were achieved with LLM-derived features (0.89, 95% CI: 0.83,0.95) and ICD+LLM diagnosis (0.88, 95% CI: 0.81,0.95). In the independent test set, ICD codes alone showed a sensitivity of 0.86 (95% CI: 0.75,0.97), PPV of 0.97 (95% CI: 0.91,1.0), and an F1 of 0.91 (95% CI: 0.84,1.0). Combining ICD and LLM-assigned diagnosis yielded a 3-point improvement in F1 score (95% CI: -0.01,0.07; p=0.2) compared to ICD alone. This combination method significantly improved sensitivity (0.92 [0.83,1.00]; p=0.008) and F1 score (0.94 [0.89,1.00]; p=0.047) relative to LLM-assigned diagnosis.
Discussion: Combining the LLM-assigned diagnosis with a single diagnostic code reduced false negatives and modestly improved the F1 score compared to either method alone, suggesting a scalable approach for improving EoE case identification in real-world data.
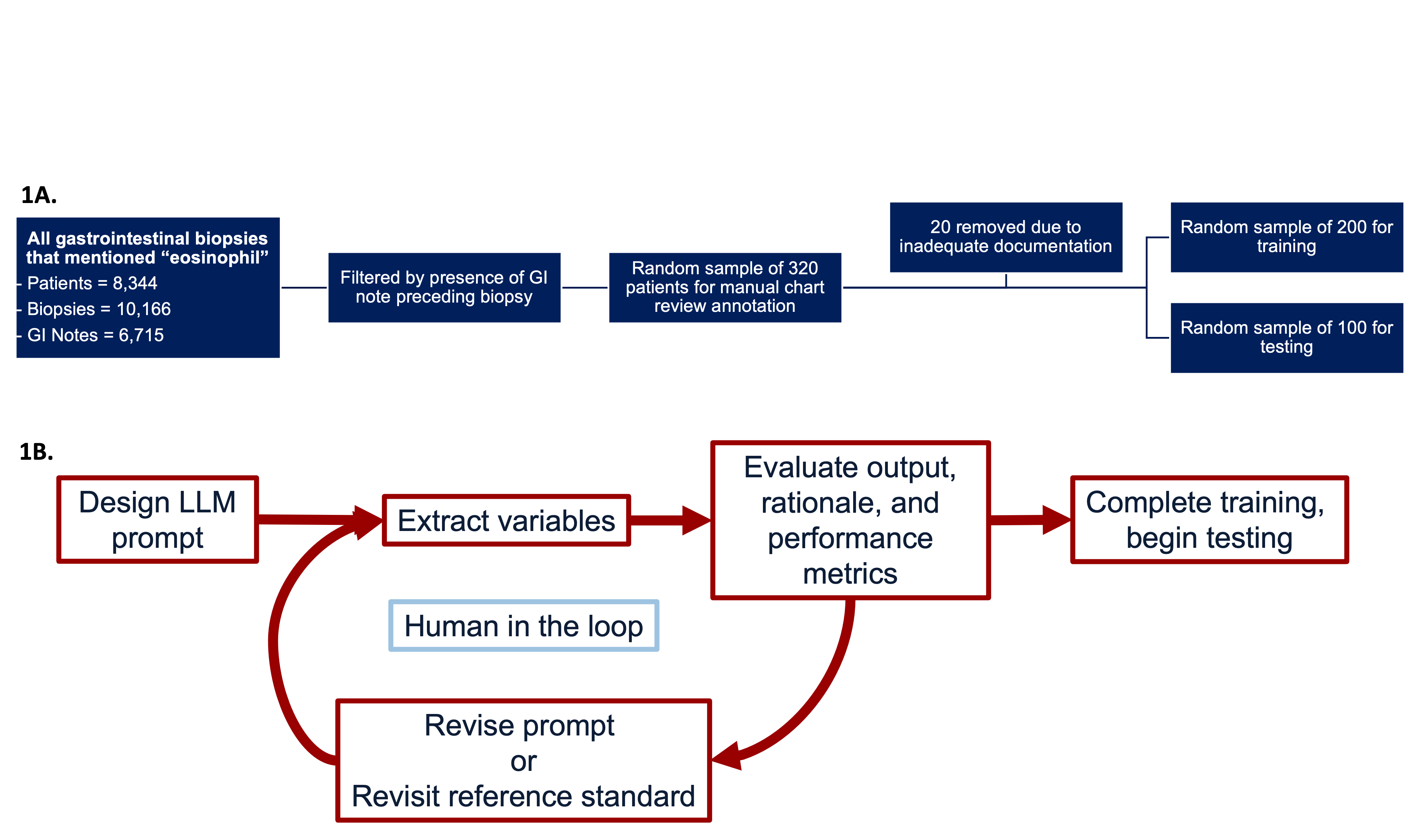
Figure: Figure (1A). Gastrointestinal biopsies between [2008-2023] that mentioned the word “eosinophil” in the pathology report were identified. The cohort was filtered to include only patients with a GI note prior to the corresponding biopsy. From this filtered cohort, a random sample of 320 patients were selected for annotation with 20 removed for inadequate documentation. Next, 200 patients were randomly allocated for training and 100 patients for testing. All selected cases underwent structured manual review to establish reference standard diagnoses. (1B) Iterative process for developing and evaluating a large language model (LLM) pipeline to extract clinical variables from unstructured text.

Figure: Table 1. Performance metrics of methods for identifying EoE in training and independent test set. ICD diagnosis refers to identification based solely on the presence of a diagnostic code. LLM-derived diagnostic features incorporate a combination of histologic findings and symptom-based criteria extracted by a large language model (LLM). LLM-assigned diagnosis reflects the LLM’s direct interpretation and assignment of an EoE diagnosis based on the provided clinical information. ICD+LLM diagnosis represents a combined approach requiring either the presence of the ICD code or an LLM-assigned diagnosis.
Disclosures:
Corey Ketchem indicated no relevant financial relationships.
Uğurcan Vurgun indicated no relevant financial relationships.
Agnes Wang indicated no relevant financial relationships.
Sunil Thomas indicated no relevant financial relationships.
Ashley Batugo indicated no relevant financial relationships.
Gary Falk indicated no relevant financial relationships.
Kristle Lynch: Medtronic – Consultant. Phathom – Consultant. Sanofi/Regeneron – Consultant. Uniquity – Consultant.
Evan Dellon: AbbVie – Consultant. Adare/Ellodi – Consultant, Grant/Research Support. Akesobio – Consultant. Alfasigma – Consultant. ALK – Consultant. Allakos – Consultant, Grant/Research Support. Amgen – Consultant. Apogee – Consultant. Apollo – Consultant. Aqilion – Consultant, Grant/Research Support. Arena/Pfizer – Consultant, Grant/Research Support. Aslan – Consultant. AstraZeneca – Consultant, Grant/Research Support. Avir – Consultant. Biocryst – Consultant. Bryn – Consultant. Calypso – Consultant. Celgene/Receptos/Bristol Myers Squibb – Consultant, Grant/Research Support. Celldex – Consultant, Grant/Research Support. Dr. Falk Pharma – Consultant. EsoCap – Consultant. Eupraxia – Consultant, Grant/Research Support. Ferring – Consultant, Grant/Research Support. GI Reviewers – Consultant. GSK – Consultant, Grant/Research Support. Holoclara – Consultant, Grant/Research Support. Invea – Consultant, Grant/Research Support. Knightpoint – Consultant. LucidDx – Consultant. Meritage – Grant/Research Support. Miraca – Grant/Research Support. Morphic – Consultant. Nexstone Immunology/Uniquity – Consultant. Nutricia – Consultant, Grant/Research Support. Parexel/Calyx – Consultant. Phathom – Consultant. Regeneron Pharmaceuticals Inc. – Consultant, Grant/Research Support. Revolo – Consultant, Grant/Research Support. Robarts/Alimentiv – Consultant. Sanofi – Consultant, Grant/Research Support. Shire/Takeda – Consultant, Grant/Research Support. Target RWE – Consultant. Third Harmonic Bio – Consultant. Uniquity – Grant/Research Support. Upstream Bio – Consultant.
Danielle Mowery: Roche – Grant/Research Support.
James Lewis: 3M – Expert witness. AbbVie – Grant/Research Support. Amgen – Advisor or Review Panel Member. Dark Canyon Laboratories – Owner/Ownership Interest. Eli Lilly – Consultant, Grant/Research Support. Johnson & Johnson – Advisory Committee/Board Member, Grant/Research Support. Odyssey Therapeutics – Advisor or Review Panel Member. Pfizer – Advisor or Review Panel Member. Sanofi – Advisor or Review Panel Member. Spyre Therapeutics – Advisor or Review Panel Member.
Corey J. Ketchem, MD1, Uğurcan Vurgun, PhD, MA2, Agnes Wang, MS3, Sunil Thomas, MBA4, Ashley Batugo, BS4, Gary Falk, MD, MS5, Kristle L.. Lynch, MD6, Evan S. Dellon, MD, MPH7, Danielle Mowery, PhD, MS8, James Lewis, MD, MSCE9. P2739 - Large Language Models Combined With a Single Diagnostic Code Detect Eosinophilic Esophagitis in Electronic Health Records With High Accuracy, ACG 2025 Annual Scientific Meeting Abstracts. Phoenix, AZ: American College of Gastroenterology.
1University of Pennsylvania Health System, Philadelphia, PA; 2University of Pennsylvania, Mill Creek, WA; 3Univeristy of Pennsylvania, Philadelphia, PA; 4University of Pennsylvania, Philadelphia, PA; 5Division of Gastroenterology and Hepatology, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA; 6The University of Pennsylvania, Philadelphia, PA; 7Center for Esophageal Diseases and Swallowing, University of North Carolina School of Medicine, Chapel Hill, NC; 8Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA; 9University of Penn, Philadelphia, PA
Introduction: Large-scale epidemiologic research for eosinophilic esophagitis (EoE) is hampered by variable accuracy of International Classification of Diseases (ICD) codes and case identification algorithms. We aimed to develop and validate a natural language processing (NLP) pipeline using large language models (LLMs) to identify EoE features and diagnosis from unstructured text, comparing performance with ICD codes and ICD+LLM combination.
Methods: We identified gastrointestinal pathology reports with any mention of “eosinophil” paired with preceding GI clinic notes (Figure 1A). 300 randomly selected patients were divided into training (n=200, 56 with EoE) and testing (n=100, 36 with EoE) sets. Manual chart review was used to assign an EoE reference standard. LLM prompt development used a human-in-the-loop approach with iterative refinements (Figure 1B). Training concluded once the LLM-assigned diagnosis exceeded the F1 score (harmonic mean of precision and recall) of ICD codes. Using the test set, we compared performance of ICD codes, LLM-derived diagnostic features, LLM-assigned diagnosis, and a combined LLM+ICD. Performance metrics included sensitivity (recall), PPV (precision), and F1 score. Nonparametric bootstrap resampling (1,000 replicates) was used to estimate 95% confidence intervals with statistical significance based on whether they excluded zero.
Results: In the training set, LLM-derived diagnostic features demonstrated the highest sensitivity (0.98, 95% CI: 0.95,1.00), while LLM-assigned diagnosis had the highest PPV (0.92, 95% CI: 0.84-1.0) and specificity (0.97, 95% CI: 0.95,1.00]) (Table 1). Comparably high F1 scores were achieved with LLM-derived features (0.89, 95% CI: 0.83,0.95) and ICD+LLM diagnosis (0.88, 95% CI: 0.81,0.95). In the independent test set, ICD codes alone showed a sensitivity of 0.86 (95% CI: 0.75,0.97), PPV of 0.97 (95% CI: 0.91,1.0), and an F1 of 0.91 (95% CI: 0.84,1.0). Combining ICD and LLM-assigned diagnosis yielded a 3-point improvement in F1 score (95% CI: -0.01,0.07; p=0.2) compared to ICD alone. This combination method significantly improved sensitivity (0.92 [0.83,1.00]; p=0.008) and F1 score (0.94 [0.89,1.00]; p=0.047) relative to LLM-assigned diagnosis.
Discussion: Combining the LLM-assigned diagnosis with a single diagnostic code reduced false negatives and modestly improved the F1 score compared to either method alone, suggesting a scalable approach for improving EoE case identification in real-world data.

Figure: Figure (1A). Gastrointestinal biopsies between [2008-2023] that mentioned the word “eosinophil” in the pathology report were identified. The cohort was filtered to include only patients with a GI note prior to the corresponding biopsy. From this filtered cohort, a random sample of 320 patients were selected for annotation with 20 removed for inadequate documentation. Next, 200 patients were randomly allocated for training and 100 patients for testing. All selected cases underwent structured manual review to establish reference standard diagnoses. (1B) Iterative process for developing and evaluating a large language model (LLM) pipeline to extract clinical variables from unstructured text.

Figure: Table 1. Performance metrics of methods for identifying EoE in training and independent test set. ICD diagnosis refers to identification based solely on the presence of a diagnostic code. LLM-derived diagnostic features incorporate a combination of histologic findings and symptom-based criteria extracted by a large language model (LLM). LLM-assigned diagnosis reflects the LLM’s direct interpretation and assignment of an EoE diagnosis based on the provided clinical information. ICD+LLM diagnosis represents a combined approach requiring either the presence of the ICD code or an LLM-assigned diagnosis.
Disclosures:
Corey Ketchem indicated no relevant financial relationships.
Uğurcan Vurgun indicated no relevant financial relationships.
Agnes Wang indicated no relevant financial relationships.
Sunil Thomas indicated no relevant financial relationships.
Ashley Batugo indicated no relevant financial relationships.
Gary Falk indicated no relevant financial relationships.
Kristle Lynch: Medtronic – Consultant. Phathom – Consultant. Sanofi/Regeneron – Consultant. Uniquity – Consultant.
Evan Dellon: AbbVie – Consultant. Adare/Ellodi – Consultant, Grant/Research Support. Akesobio – Consultant. Alfasigma – Consultant. ALK – Consultant. Allakos – Consultant, Grant/Research Support. Amgen – Consultant. Apogee – Consultant. Apollo – Consultant. Aqilion – Consultant, Grant/Research Support. Arena/Pfizer – Consultant, Grant/Research Support. Aslan – Consultant. AstraZeneca – Consultant, Grant/Research Support. Avir – Consultant. Biocryst – Consultant. Bryn – Consultant. Calypso – Consultant. Celgene/Receptos/Bristol Myers Squibb – Consultant, Grant/Research Support. Celldex – Consultant, Grant/Research Support. Dr. Falk Pharma – Consultant. EsoCap – Consultant. Eupraxia – Consultant, Grant/Research Support. Ferring – Consultant, Grant/Research Support. GI Reviewers – Consultant. GSK – Consultant, Grant/Research Support. Holoclara – Consultant, Grant/Research Support. Invea – Consultant, Grant/Research Support. Knightpoint – Consultant. LucidDx – Consultant. Meritage – Grant/Research Support. Miraca – Grant/Research Support. Morphic – Consultant. Nexstone Immunology/Uniquity – Consultant. Nutricia – Consultant, Grant/Research Support. Parexel/Calyx – Consultant. Phathom – Consultant. Regeneron Pharmaceuticals Inc. – Consultant, Grant/Research Support. Revolo – Consultant, Grant/Research Support. Robarts/Alimentiv – Consultant. Sanofi – Consultant, Grant/Research Support. Shire/Takeda – Consultant, Grant/Research Support. Target RWE – Consultant. Third Harmonic Bio – Consultant. Uniquity – Grant/Research Support. Upstream Bio – Consultant.
Danielle Mowery: Roche – Grant/Research Support.
James Lewis: 3M – Expert witness. AbbVie – Grant/Research Support. Amgen – Advisor or Review Panel Member. Dark Canyon Laboratories – Owner/Ownership Interest. Eli Lilly – Consultant, Grant/Research Support. Johnson & Johnson – Advisory Committee/Board Member, Grant/Research Support. Odyssey Therapeutics – Advisor or Review Panel Member. Pfizer – Advisor or Review Panel Member. Sanofi – Advisor or Review Panel Member. Spyre Therapeutics – Advisor or Review Panel Member.
Corey J. Ketchem, MD1, Uğurcan Vurgun, PhD, MA2, Agnes Wang, MS3, Sunil Thomas, MBA4, Ashley Batugo, BS4, Gary Falk, MD, MS5, Kristle L.. Lynch, MD6, Evan S. Dellon, MD, MPH7, Danielle Mowery, PhD, MS8, James Lewis, MD, MSCE9. P2739 - Large Language Models Combined With a Single Diagnostic Code Detect Eosinophilic Esophagitis in Electronic Health Records With High Accuracy, ACG 2025 Annual Scientific Meeting Abstracts. Phoenix, AZ: American College of Gastroenterology.
