Monday Poster Session
Category: Esophagus
P2763 - Automated Detection of Barrett's Esophagus Dysplasia Using Deep Learning With Pathology Foundation Models on a Large Multicenter Cohort of Whole Slide Images
Monday, October 27, 2025
10:30 AM - 4:00 PM PDT
Location: Exhibit Hall

Don C. Codipilly, MD (he/him/his)
Mayo Clinic
Rochester, MN
Presenting Author(s)
Award: ACG Outstanding Research Award in the Esophagus Category
Award: ACG Presidential Poster Award
Don C. Codipilly, MD1, Amirali Khosravi, MD1, Jaidip Jagtap, PhD1, Jason Lewis, MD2, Catherine Hagen, MD1, Shajan Peter, MD3, Vani Konda, MD, FACG4, Fouad Otaki, MD5, Sachin Wani, MD6, Nicholas J. Shaheen, MD, MPH, MACG7, David Katzka, MD8, Bradley Erickson, MD, PhD1, Prasad Iyer, MD, MS9
1Mayo Clinic, Rochester, MN; 2Mayo Clinic, Jacksonville, FL; 3University of Alabama at Birmingham, Birmingham, AL; 4Baylor Scott & White Medical Center, Dallas, TX; 5Oregon Health & Science University, Portland, OR; 6University of Colorado-Anschutz Campus, Aurora, CO; 7University of North Carolina at Chapel Hill, Chapel Hill, NC; 8Columbia University, New York City, NY; 9Mayo Clinic, Phoenix, AZ
Introduction: The histologic diagnosis of dysplastic Barrett's esophagus (BE) suffers from substantial interobserver variability due to subjective criteria and inflammatory artifacts, leading to dysplastic overcalls particularly in the community setting. Precise dysplasia grading is critical for determining appropriate management. We aimed to develop and validate a deep learning model using a large multi-center cohort to improve the accuracy of BE dysplasia grade diagnosis.
Methods: We utilized 969 whole slide images (WSIs) from four academic centers (Mayo Clinic, Baylor, OHSU, UAB) collected from 1992 to 2022. All patients had endoscopic BE (≥1 cm columnar mucosa) with confirmed intestinal metaplasia. Ground truth was established by consensus review by two expert GI pathologists. Data were stratified by dysplasia grade and center and split 70-15-15 for training, validation, and testing. We employed a semi-supervised approach using Clustering-constrained Attention Multiple Instance Learning (CLAM) architecture. WSIs were divided into patches with features extracted using the Midnight foundation model, a pre-trained deep learning model that learned to identify histologic patterns from large datasets of pathology images (Figure 1). Three binary classifiers identified candidate patches followed by the final CLAM model predicting WSI-level dysplasia grade. Model performance was evaluated using AUC and F1 score, a balanced measure of diagnostic accuracy accounting for both correct identification of dysplasia and avoidance of false positives.
Results: The mean age was 64.6 ± 12.8 years and 81.9% were men. Dysplasia grade by consensus diagnosis was 365 nondysplastic BE (NDBE), 319 LGD, and 285 high-grade dysplasia (HGD). On the standalone test set, the model achieved AUC of 95%, F1 score of 82%, sensitivity of 82%, and specificity of 91% for three-class grading (Table 1). For binary classification of any dysplasia versus NDBE, AUC was 97%, F1 score was 93%, sensitivity of 90%, and specificity of 94%.
Discussion: Our model demonstrates promising performance for BE dysplasia diagnosis grade. The combination of a large multi-center training dataset and foundation model training provides inherent robustness accounting for staining variations across institutions without requiring center-specific adaptations. This approach may serve as a valuable adjunct to pathologist review, improving diagnostic consistency. External validation of this model is planned.
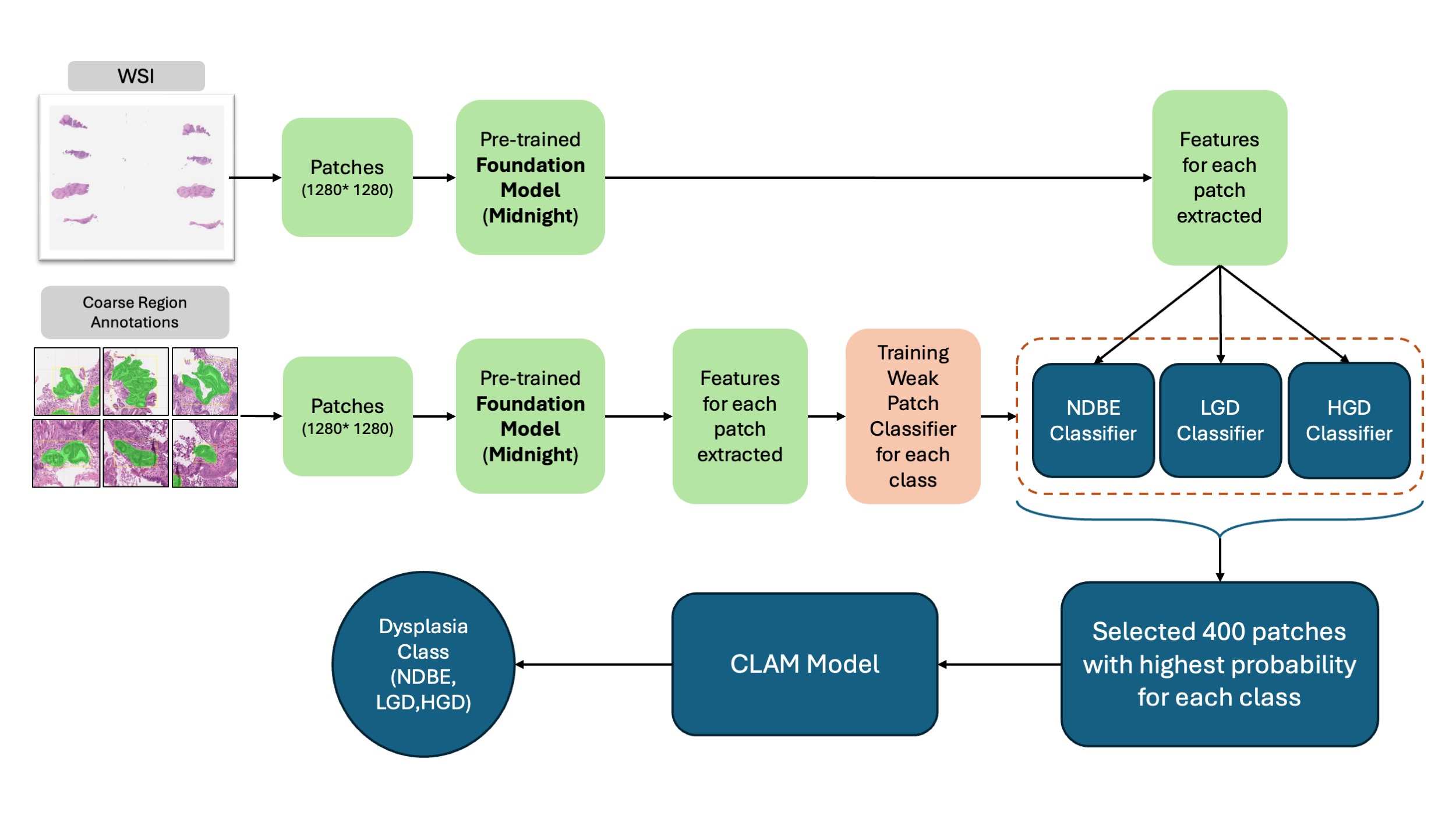
Figure: Figure 1. Pre-processing, Training, and Validation of the Deep Learning Model
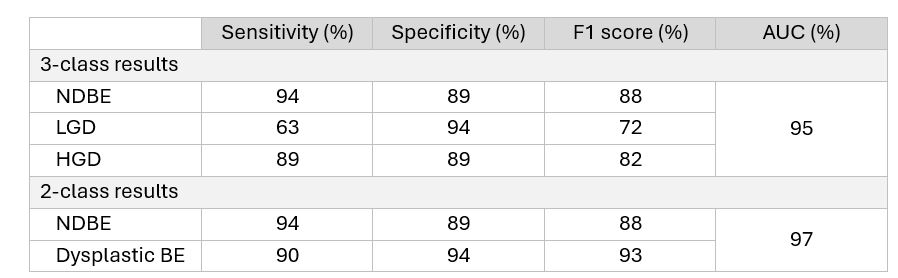
Figure: Table 1. Performance of Deep Learning Model on the Diagnosis of Dysplasia Grade, Compared to Expert GI Pathologist Consensus
Disclosures:
Don Codipilly indicated no relevant financial relationships.
Amirali Khosravi indicated no relevant financial relationships.
Jaidip Jagtap indicated no relevant financial relationships.
Jason Lewis indicated no relevant financial relationships.
Catherine Hagen indicated no relevant financial relationships.
Shajan Peter: Castle biosciences – Advisory Committee/Board Member. Olympus corporation – Advisory Committee/Board Member.
Vani Konda: Ambu – Consultant. Braintree – Consultant. Castle Biosciences – Consultant. Exact – Consultant. Medtronic – Consultant. Pentax – Consultant.
Fouad Otaki indicated no relevant financial relationships.
Sachin Wani: CDx – Grant/Research Support. Exact – Consultant, Grant/Research Support. Lucid Diagnostics – Grant/Research Support. Previse – Consultant.
Nicholas Shaheen: Aqua – Grant/Research Support. CDx – Grant/Research Support. Cook Medical – Consultant. GIE Medical – Grant/Research Support. Interpace Diagnostics – Grant/Research Support. Lucid Diagnostics – Grant/Research Support. Medtronic – Grant/Research Support. Pentax – Grant/Research Support. Steris – Grant/Research Support.
David Katzka indicated no relevant financial relationships.
Bradley Erickson: FlowSigma, INC – Officer. Yunu, INC – Officer.
Prasad Iyer: CDx medical – Consultant, Grant/Research Support. Exact – Grant/Research Support. Exact Sciences – Consultant, Grant/Research Support. Medtronic – Consultant. Pentax Medical – Consultant, Grant/Research Support.
Don C. Codipilly, MD1, Amirali Khosravi, MD1, Jaidip Jagtap, PhD1, Jason Lewis, MD2, Catherine Hagen, MD1, Shajan Peter, MD3, Vani Konda, MD, FACG4, Fouad Otaki, MD5, Sachin Wani, MD6, Nicholas J. Shaheen, MD, MPH, MACG7, David Katzka, MD8, Bradley Erickson, MD, PhD1, Prasad Iyer, MD, MS9. P2763 - Automated Detection of Barrett's Esophagus Dysplasia Using Deep Learning With Pathology Foundation Models on a Large Multicenter Cohort of Whole Slide Images, ACG 2025 Annual Scientific Meeting Abstracts. Phoenix, AZ: American College of Gastroenterology.
Award: ACG Presidential Poster Award
Don C. Codipilly, MD1, Amirali Khosravi, MD1, Jaidip Jagtap, PhD1, Jason Lewis, MD2, Catherine Hagen, MD1, Shajan Peter, MD3, Vani Konda, MD, FACG4, Fouad Otaki, MD5, Sachin Wani, MD6, Nicholas J. Shaheen, MD, MPH, MACG7, David Katzka, MD8, Bradley Erickson, MD, PhD1, Prasad Iyer, MD, MS9
1Mayo Clinic, Rochester, MN; 2Mayo Clinic, Jacksonville, FL; 3University of Alabama at Birmingham, Birmingham, AL; 4Baylor Scott & White Medical Center, Dallas, TX; 5Oregon Health & Science University, Portland, OR; 6University of Colorado-Anschutz Campus, Aurora, CO; 7University of North Carolina at Chapel Hill, Chapel Hill, NC; 8Columbia University, New York City, NY; 9Mayo Clinic, Phoenix, AZ
Introduction: The histologic diagnosis of dysplastic Barrett's esophagus (BE) suffers from substantial interobserver variability due to subjective criteria and inflammatory artifacts, leading to dysplastic overcalls particularly in the community setting. Precise dysplasia grading is critical for determining appropriate management. We aimed to develop and validate a deep learning model using a large multi-center cohort to improve the accuracy of BE dysplasia grade diagnosis.
Methods: We utilized 969 whole slide images (WSIs) from four academic centers (Mayo Clinic, Baylor, OHSU, UAB) collected from 1992 to 2022. All patients had endoscopic BE (≥1 cm columnar mucosa) with confirmed intestinal metaplasia. Ground truth was established by consensus review by two expert GI pathologists. Data were stratified by dysplasia grade and center and split 70-15-15 for training, validation, and testing. We employed a semi-supervised approach using Clustering-constrained Attention Multiple Instance Learning (CLAM) architecture. WSIs were divided into patches with features extracted using the Midnight foundation model, a pre-trained deep learning model that learned to identify histologic patterns from large datasets of pathology images (Figure 1). Three binary classifiers identified candidate patches followed by the final CLAM model predicting WSI-level dysplasia grade. Model performance was evaluated using AUC and F1 score, a balanced measure of diagnostic accuracy accounting for both correct identification of dysplasia and avoidance of false positives.
Results: The mean age was 64.6 ± 12.8 years and 81.9% were men. Dysplasia grade by consensus diagnosis was 365 nondysplastic BE (NDBE), 319 LGD, and 285 high-grade dysplasia (HGD). On the standalone test set, the model achieved AUC of 95%, F1 score of 82%, sensitivity of 82%, and specificity of 91% for three-class grading (Table 1). For binary classification of any dysplasia versus NDBE, AUC was 97%, F1 score was 93%, sensitivity of 90%, and specificity of 94%.
Discussion: Our model demonstrates promising performance for BE dysplasia diagnosis grade. The combination of a large multi-center training dataset and foundation model training provides inherent robustness accounting for staining variations across institutions without requiring center-specific adaptations. This approach may serve as a valuable adjunct to pathologist review, improving diagnostic consistency. External validation of this model is planned.

Figure: Figure 1. Pre-processing, Training, and Validation of the Deep Learning Model

Figure: Table 1. Performance of Deep Learning Model on the Diagnosis of Dysplasia Grade, Compared to Expert GI Pathologist Consensus
Disclosures:
Don Codipilly indicated no relevant financial relationships.
Amirali Khosravi indicated no relevant financial relationships.
Jaidip Jagtap indicated no relevant financial relationships.
Jason Lewis indicated no relevant financial relationships.
Catherine Hagen indicated no relevant financial relationships.
Shajan Peter: Castle biosciences – Advisory Committee/Board Member. Olympus corporation – Advisory Committee/Board Member.
Vani Konda: Ambu – Consultant. Braintree – Consultant. Castle Biosciences – Consultant. Exact – Consultant. Medtronic – Consultant. Pentax – Consultant.
Fouad Otaki indicated no relevant financial relationships.
Sachin Wani: CDx – Grant/Research Support. Exact – Consultant, Grant/Research Support. Lucid Diagnostics – Grant/Research Support. Previse – Consultant.
Nicholas Shaheen: Aqua – Grant/Research Support. CDx – Grant/Research Support. Cook Medical – Consultant. GIE Medical – Grant/Research Support. Interpace Diagnostics – Grant/Research Support. Lucid Diagnostics – Grant/Research Support. Medtronic – Grant/Research Support. Pentax – Grant/Research Support. Steris – Grant/Research Support.
David Katzka indicated no relevant financial relationships.
Bradley Erickson: FlowSigma, INC – Officer. Yunu, INC – Officer.
Prasad Iyer: CDx medical – Consultant, Grant/Research Support. Exact – Grant/Research Support. Exact Sciences – Consultant, Grant/Research Support. Medtronic – Consultant. Pentax Medical – Consultant, Grant/Research Support.
Don C. Codipilly, MD1, Amirali Khosravi, MD1, Jaidip Jagtap, PhD1, Jason Lewis, MD2, Catherine Hagen, MD1, Shajan Peter, MD3, Vani Konda, MD, FACG4, Fouad Otaki, MD5, Sachin Wani, MD6, Nicholas J. Shaheen, MD, MPH, MACG7, David Katzka, MD8, Bradley Erickson, MD, PhD1, Prasad Iyer, MD, MS9. P2763 - Automated Detection of Barrett's Esophagus Dysplasia Using Deep Learning With Pathology Foundation Models on a Large Multicenter Cohort of Whole Slide Images, ACG 2025 Annual Scientific Meeting Abstracts. Phoenix, AZ: American College of Gastroenterology.


