Sunday Poster Session
Category: IBD
P1156 - Integrative Transcription Factor-Gene-Metabolite Network Analysis Reveals Mitochondrial Regulatory Nodes in Crohn’s Disease
Sunday, October 26, 2025
3:30 PM - 7:00 PM PDT
Location: Exhibit Hall
- KB
Kiandokht Bashiri, MD, MSc
MedStar Georgetown University Hospital
Washington, DC
Presenting Author(s)
Kiandokht Bashiri, MD, MSc1, Alireza Meighani, MD1, Reezwana Chowdhury, MD2, Angela Wu, MD1, Mark Mattar, MD1
1MedStar Georgetown University Hospital, Washington, DC; 2Johns Hopkins University, Baltimore, MD
Introduction: Mitochondrial dysfunction is a well-recognized hallmark of Crohn’s disease (CD), yet its upstream transcriptional regulation and downstream metabolic consequences remain poorly characterized. We constructed a triadic transcription factor (TF) → gene → metabolite network to uncover regulatory axes underlying mitochondrial alterations in CD.
Methods: Using publicly available whole blood transcriptomic data (GSE119600, Ostrowski et al., 2019), we identified differentially expressed genes (DEGs) in CD compared to controls. Upstream TFs predicted to regulate these DEGs were identified via ChEA3. Independently, stool metabolomics data (ST000923, Avila-Pacheco et al., 2017) were screened for mitochondrial-related metabolites. A directed multi-layer network was constructed by linking TFs to their target genes and genes to biochemically associated metabolites, with emphasis on mitochondrial pathways.
Results: The resulting TF–gene–metabolite network highlighted mitochondrial genes such as COX5A, NDUFB8, and NDUFS4, and metabolites including lactate, fumarate, and NADH, all known to be dysregulated in CD. Notably, predicted regulators such as MTERF3 and SNAPC5, along with lesser-studied TFs like CENPX and GTF3A, were integrated into the network, suggesting novel transcriptional control points. Additionally, the inclusion of uncommon metabolites (e.g., 5-acetylamino-6-amino-3-methyluracil) expands the landscape of potentially relevant mitochondrial disruptions.
Discussion: This integrative tri-layered network approach underscores the pivotal role of mitochondrial dysfunction in CD and highlights potential regulatory nodes that bridge transcriptional and metabolic dysregulation. While several interactions align with prior findings, others-particularly those involving understudied TFs and rare metabolites-may indicate uncharacterized pathways, necessitating further experimental validation.
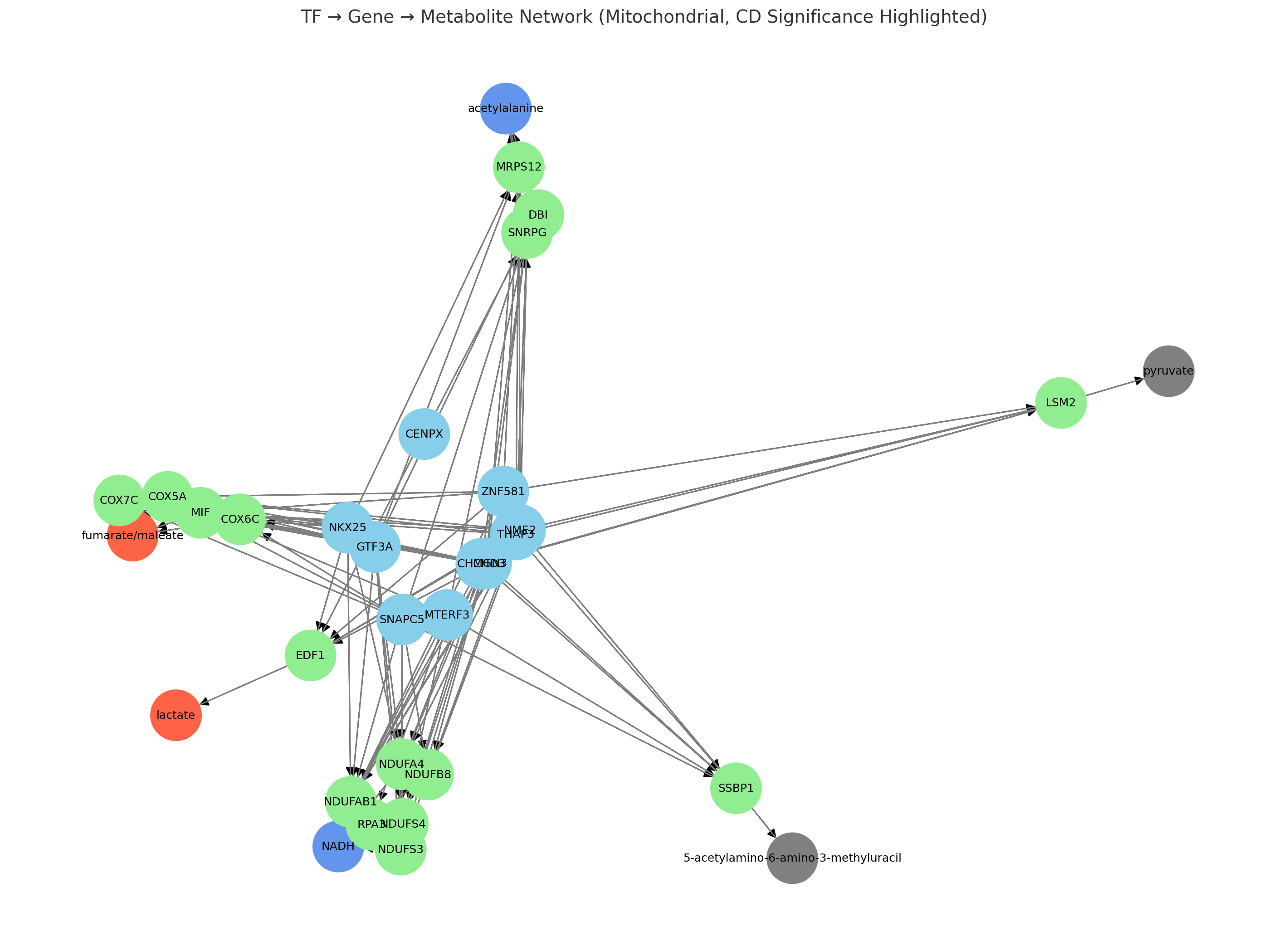
Figure: Integrative transcription factor-gene-metabolite network highlighting potential dysregulated mitochondrial pathways in Crohn’s disease; nodes represent transcription factors (blue), target genes (green), upregulated CD-altered metabolites (red), downregulated metabolites (blue), and other metabolites (gray), with edges indicating regulatory (TF → Gene) and metabolic (Gene → Metabolite) interactions.
Disclosures:
Kiandokht Bashiri indicated no relevant financial relationships.
Alireza Meighani indicated no relevant financial relationships.
Reezwana Chowdhury indicated no relevant financial relationships.
Angela Wu indicated no relevant financial relationships.
Mark Mattar: AbbVie – Consultant, Speakers Bureau. Celltrion – Speakers Bureau. Johnson & Johnson – Consultant, Speakers Bureau.
Kiandokht Bashiri, MD, MSc1, Alireza Meighani, MD1, Reezwana Chowdhury, MD2, Angela Wu, MD1, Mark Mattar, MD1. P1156 - Integrative Transcription Factor-Gene-Metabolite Network Analysis Reveals Mitochondrial Regulatory Nodes in Crohn’s Disease, ACG 2025 Annual Scientific Meeting Abstracts. Phoenix, AZ: American College of Gastroenterology.
1MedStar Georgetown University Hospital, Washington, DC; 2Johns Hopkins University, Baltimore, MD
Introduction: Mitochondrial dysfunction is a well-recognized hallmark of Crohn’s disease (CD), yet its upstream transcriptional regulation and downstream metabolic consequences remain poorly characterized. We constructed a triadic transcription factor (TF) → gene → metabolite network to uncover regulatory axes underlying mitochondrial alterations in CD.
Methods: Using publicly available whole blood transcriptomic data (GSE119600, Ostrowski et al., 2019), we identified differentially expressed genes (DEGs) in CD compared to controls. Upstream TFs predicted to regulate these DEGs were identified via ChEA3. Independently, stool metabolomics data (ST000923, Avila-Pacheco et al., 2017) were screened for mitochondrial-related metabolites. A directed multi-layer network was constructed by linking TFs to their target genes and genes to biochemically associated metabolites, with emphasis on mitochondrial pathways.
Results: The resulting TF–gene–metabolite network highlighted mitochondrial genes such as COX5A, NDUFB8, and NDUFS4, and metabolites including lactate, fumarate, and NADH, all known to be dysregulated in CD. Notably, predicted regulators such as MTERF3 and SNAPC5, along with lesser-studied TFs like CENPX and GTF3A, were integrated into the network, suggesting novel transcriptional control points. Additionally, the inclusion of uncommon metabolites (e.g., 5-acetylamino-6-amino-3-methyluracil) expands the landscape of potentially relevant mitochondrial disruptions.
Discussion: This integrative tri-layered network approach underscores the pivotal role of mitochondrial dysfunction in CD and highlights potential regulatory nodes that bridge transcriptional and metabolic dysregulation. While several interactions align with prior findings, others-particularly those involving understudied TFs and rare metabolites-may indicate uncharacterized pathways, necessitating further experimental validation.

Figure: Integrative transcription factor-gene-metabolite network highlighting potential dysregulated mitochondrial pathways in Crohn’s disease; nodes represent transcription factors (blue), target genes (green), upregulated CD-altered metabolites (red), downregulated metabolites (blue), and other metabolites (gray), with edges indicating regulatory (TF → Gene) and metabolic (Gene → Metabolite) interactions.
Disclosures:
Kiandokht Bashiri indicated no relevant financial relationships.
Alireza Meighani indicated no relevant financial relationships.
Reezwana Chowdhury indicated no relevant financial relationships.
Angela Wu indicated no relevant financial relationships.
Mark Mattar: AbbVie – Consultant, Speakers Bureau. Celltrion – Speakers Bureau. Johnson & Johnson – Consultant, Speakers Bureau.
Kiandokht Bashiri, MD, MSc1, Alireza Meighani, MD1, Reezwana Chowdhury, MD2, Angela Wu, MD1, Mark Mattar, MD1. P1156 - Integrative Transcription Factor-Gene-Metabolite Network Analysis Reveals Mitochondrial Regulatory Nodes in Crohn’s Disease, ACG 2025 Annual Scientific Meeting Abstracts. Phoenix, AZ: American College of Gastroenterology.
